Regulation of transcription by SMU.1349, a TetR family regulator, in Streptococcus mutans
- PMID: 21965566
- PMCID: PMC3232899
- DOI: 10.1128/JB.06122-11
Regulation of transcription by SMU.1349, a TetR family regulator, in Streptococcus mutans
Abstract
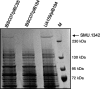
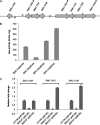
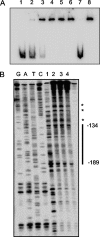
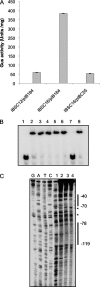
The TetR family of transcriptional regulators is ubiquitous in bacteria, where it plays an important role in bacterial gene expression. Streptococcus mutans, a gram-positive pathogen considered to be the primary etiological agent in the formation of dental caries, encodes at least 18 TetR regulators. Here we characterized one such TetR regulator, SMU.1349, encoded by the TnSmu2 operon, which appeared to be acquired by the organism via horizontal gene transfer. SMU.1349 is transcribed divergently from the rest of the genes encoded by the operon. By the use of a transcriptional reporter system and semiquantitative reverse transcription-PCR (RT-PCR), we demonstrated that SMU.1349 activates the transcription of several genes that are encoded within the TnSmu2 operon. Gel mobility shift and DNase I footprinting assays with purified SMU.1349 protein demonstrated binding to the intergenic region between SMU.1349 and the TnSmu2 operon; therefore, SMU.1349 is directly involved in gene transcription. Using purified S. mutans RpoD and Escherichia coli RNA polymerase, we also demonstrated in an in vitro transcription assay that SMU.1349 could activate transcription from the TnSmu2 operon promoter. Furthermore, we showed that SMU.1349 could also repress transcription from its own promoter by binding to the intergenic region, suggesting that SMU.1349 acts as both an activator and a repressor. Thus, unlike most of the TetR family proteins, which generally function as transcriptional repressors, SMU.1349 is unique in that it can function as both.
Figures

References
-
- Aravind L., Anantharaman V., Balaji S., Babu M. M., Iyer L. M. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231–262 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

