Qualitative and quantitative cellular glycomics of glycosphingolipids based on rhodococcal endoglycosylceramidase-assisted glycan cleavage, glycoblotting-assisted sample preparation, and matrix-assisted laser desorption ionization tandem time-of-flight mass spectrometry analysis
- PMID: 21965662
- PMCID: PMC3308876
- DOI: 10.1074/jbc.M111.301796
Qualitative and quantitative cellular glycomics of glycosphingolipids based on rhodococcal endoglycosylceramidase-assisted glycan cleavage, glycoblotting-assisted sample preparation, and matrix-assisted laser desorption ionization tandem time-of-flight mass spectrometry analysis
Abstract
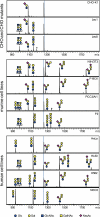
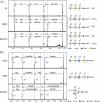
Glycosphingolipids (GSLs) are crucially important components of the cellular membrane, where they comprise microdomains with many critical biological functions. Despite this fact, qualitative and quantitative techniques for the analysis of GSLs still lag behind the needs of researchers. In this study, a reliable procedure for the elucidation of cellular GSL-glycomes was established based on (a) enzymatic glycan cleavage by endoglycosylceramidases derived from Rhodococcus sp. in combination with (b) glycoblotting-assisted sample preparation. The mixture of endoglycosylceramidase I and II was employed to maximize the release of glycan moieties from the major classes of GSLs (i.e. ganglio-, (neo)lacto- and globo-series GSLs). The glycoblotting technique enabled the quantitative detection of GSL-glycans using as few as 2 × 10(5) cells. Thirty-seven different kinds of cellular GSL glycans were successfully observed in 11 kinds of cells, including Chinese hamster ovary cells and their lectin-resistant mutants as well as murine and human embryonic carcinoma cells. Furthermore, in-depth structural clarification in terms of discrimination of isomers was achieved by MALDI-TOF/TOF mass spectrometry analysis and/or linkage-specific glycosidase digestion. These novel analytical techniques were shown to be capable of delineating cell-specific GSL-glycomes. Thus, they are anticipated to have a broad range of applications for the characterization, description, and comparison of various cellular/tissue samples in the fields of drug discovery and regenerative medicine.
Figures



References
-
- Kannagi R., Levery S. B., Ishigami F., Hakomori S., Shevinsky L. H., Knowles B. B., Solter D. (1983) J. Biol. Chem. 258, 8934–8942 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

