Special AT-rich binding protein-2 (SATB2) differentially affects disease-causing p63 mutant proteins
- PMID: 21965674
- PMCID: PMC3220488
- DOI: 10.1074/jbc.M111.271189
Special AT-rich binding protein-2 (SATB2) differentially affects disease-causing p63 mutant proteins
Abstract
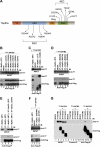
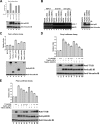
p63, a p53 family member, is critical for proper skin and limb development and directly regulates gene expression in the ectoderm. Mice lacking p63 exhibit skin and craniofacial defects including cleft palate. In humans p63 mutations are associated with several distinct developmental syndromes. p63 sterile-α-motif domain, AEC (ankyloblepharon-ectodermal dysplasia-clefting)-associated mutations are associated with a high prevalence of orofacial clefting disorders, which are less common in EEC (ectrodactyly-ectodermal dysplasia-clefting) patients with DNA binding domain p63 mutations. However, the mechanisms by which these mutations differentially influence p63 function remain unclear, and interactions with other proteins implicated in craniofacial development have not been identified. Here, we show that AEC p63 mutations affect the ability of the p63 protein to interact with special AT-rich binding protein-2 (SATB2), which has recently also been implicated in the development of cleft palate. p63 and SATB2 are co-expressed early in development in the ectoderm of the first and second branchial arches, two essential sites where signaling is required for craniofacial patterning. SATB2 attenuates p63-mediated gene expression of perp (p53 apoptosis effector related to PMP-22), a critical downstream target gene during development, and specifically decreases p63 perp promoter binding. Interestingly, AEC but not EEC p63 mutations affect the ability of p63 to interact with SATB2 and the inhibitory effects of SATB2 on p63 transactivation of perp are most pronounced for AEC-associated p63 mutations. Our findings reveal a novel gain-of-function property of AEC-causing p63 mutations and identify SATB2 as the first p63 binding partner that differentially influences AEC and EEC p63 mutant proteins.
Figures



References
-
- Melino G., De Laurenzi V., Vousden K. H. (2002) Nat. Rev. Cancer 2, 605–615 - PubMed
-
- Yang A., McKeon F. (2000) Nat. Rev. Mol. Cell Biol. 1, 199–207 - PubMed
-
- Carroll D. K., Carroll J. S., Leong C. O., Cheng F., Brown M., Mills A. A., Brugge J. S., Ellisen L. W. (2006) Nat. Cell Biol. 8, 551–561 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

