Large-scale gene-centric analysis identifies novel variants for coronary artery disease
- PMID: 21966275
- PMCID: PMC3178591
- DOI: 10.1371/journal.pgen.1002260
Large-scale gene-centric analysis identifies novel variants for coronary artery disease
Erratum in
- PLoS Genet. 2-12 Aug;8(8). doi: 10.1371/annotation/120649cf-8c28-43c9-a688-c7cd65eb1aec. Roosendaal, Frits R [corrected to Rosendaal, Frits R] doi: 10.1371/annotation/120649cf-8c28-43c9-a688-c7cd65eb1aec
Abstract
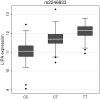
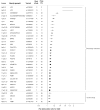
Coronary artery disease (CAD) has a significant genetic contribution that is incompletely characterized. To complement genome-wide association (GWA) studies, we conducted a large and systematic candidate gene study of CAD susceptibility, including analysis of many uncommon and functional variants. We examined 49,094 genetic variants in ∼2,100 genes of cardiovascular relevance, using a customised gene array in 15,596 CAD cases and 34,992 controls (11,202 cases and 30,733 controls of European descent; 4,394 cases and 4,259 controls of South Asian origin). We attempted to replicate putative novel associations in an additional 17,121 CAD cases and 40,473 controls. Potential mechanisms through which the novel variants could affect CAD risk were explored through association tests with vascular risk factors and gene expression. We confirmed associations of several previously known CAD susceptibility loci (eg, 9p21.3:p<10(-33); LPA:p<10(-19); 1p13.3:p<10(-17)) as well as three recently discovered loci (COL4A1/COL4A2, ZC3HC1, CYP17A1:p<5×10(-7)). However, we found essentially null results for most previously suggested CAD candidate genes. In our replication study of 24 promising common variants, we identified novel associations of variants in or near LIPA, IL5, TRIB1, and ABCG5/ABCG8, with per-allele odds ratios for CAD risk with each of the novel variants ranging from 1.06-1.09. Associations with variants at LIPA, TRIB1, and ABCG5/ABCG8 were supported by gene expression data or effects on lipid levels. Apart from the previously reported variants in LPA, none of the other ∼4,500 low frequency and functional variants showed a strong effect. Associations in South Asians did not differ appreciably from those in Europeans, except for 9p21.3 (per-allele odds ratio: 1.14 versus 1.27 respectively; P for heterogeneity = 0.003). This large-scale gene-centric analysis has identified several novel genes for CAD that relate to diverse biochemical and cellular functions and clarified the literature with regard to many previously suggested genes.
Conflict of interest statement
Muredach P Reilly and Daniel J Rader have a research grant from GlaxoSmithKline. The division of Pharmacoepidemiology and Clinical Pharmacology employing Bas Peters, Olaf Klungel, Anthonius de Boer, and Anke-Hilse Maitland-van der Zee has received unrestricted funding for pharmacoepidemiological research from GlaxoSmithKline, Novo Nordisk, the private-public funded Top Institute Pharma (www.tipharma.nl, includes co-funding from universities, government, and industry), the Dutch Medicines Evaluation Board, and the Dutch Ministry of Health. Arthur AM Wilde is a consultant for Transgenomics (Familion test) and Sorin. No other disclosures were reported.
Figures


References
-
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. - PubMed
Publication types
MeSH terms
Grants and funding
- G1000143/MRC_/Medical Research Council/United Kingdom
- MC_U106179471/MRC_/Medical Research Council/United Kingdom
- G0401527/MRC_/Medical Research Council/United Kingdom
- G0701863/MRC_/Medical Research Council/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- MC_U105260792/MRC_/Medical Research Council/United Kingdom
- RP-PG-0310-1002/DH_/Department of Health/United Kingdom
- G0801056/MRC_/Medical Research Council/United Kingdom
- G0700931/MRC_/Medical Research Council/United Kingdom
- RG/08/014/24067/BHF_/British Heart Foundation/United Kingdom
- MC_U137686857/MRC_/Medical Research Council/United Kingdom
- G0601966/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

