Role of chaperone mediated autophagy (CMA) in the degradation of misfolded N-CoR protein in non-small cell lung cancer (NSCLC) cells
- PMID: 21966475
- PMCID: PMC3179509
- DOI: 10.1371/journal.pone.0025268
Role of chaperone mediated autophagy (CMA) in the degradation of misfolded N-CoR protein in non-small cell lung cancer (NSCLC) cells
Abstract
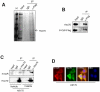
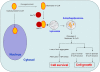
Nuclear receptor co-repressor (N-CoR) plays important role in transcriptional control mediated by several tumor suppressor proteins. Recently, we reported a role of misfolded-conformation dependent loss (MCDL) of N-CoR in the activation of oncogenic survival pathway in acute promyelocytic leukemia (APL). Since N-CoR plays important role in cellular homeostasis in various tissues, therefore, we hypothesized that an APL like MCDL of N-CoR might also be involved in other malignancy. Indeed, our initial screening of N-CoR status in various leukemia and solid tumor cells revealed an APL like MCDL of N-CoR in primary and secondary tumor cells derived from non-small cell lung cancer (NSCLC). The NSCLC cell specific N-CoR loss could be blocked by Kaletra, a clinical grade protease inhibitor and by genistein, an inhibitor of N-CoR misfolding previously characterized by us. The misfolded N-CoR presented in NSCLC cells was linked to the amplification of ER stress and was subjected to degradation by NSCLC cell specific aberrant protease activity. In NSCLC cells, misfolded N-CoR was found to be associated with Hsc70, a molecular chaperone involved in chaperone mediated autophagy (CMA). Genetic and chemical inhibition of Lamp2A, a rate limiting factor of CMA, significantly blocked the loss of N-CoR in NSCLC cells, suggesting a crucial role of CMA in N-CoR degradation. These findings identify an important role of CMA-induced degradation of misfolded N-CoR in the neutralization of ER stress and suggest a possible role of misfolded N-CoR protein in the activation of oncogenic survival pathway in NSCLC cells.
Conflict of interest statement
Figures




References
-
- Halmos B, Huettner CS, Kocher O, Ferenczi K, Karp DD, et al. Down-regulation and anti-proliferative role of C/EBPalpha in lung cancer. Cancer Res. 2002;62:528–534. - PubMed
-
- Halmos B, Bassères DS, Monti S, D'Aló F, Dayaram T, et al. A transcriptional profiling study of CCAAT/Enhancer binding protein targets identifies hepatocyte nuclear factor 3 as a novel tumor suppressor in lung cancer. Cancer Res. 2004;64:4137–4147. - PubMed
-
- Khan MM, Nomura T, Kim H, Kaul SC, Wadhwa R, et al. PML-RAR alleviates transcriptional repression mediated by tumor suppressor Rb. J Biol Chem. 2001;276:43491–43494. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

