A test of the coordinated expression hypothesis for the origin and maintenance of the GAL cluster in yeast
- PMID: 21966486
- PMCID: PMC3178652
- DOI: 10.1371/journal.pone.0025290
A test of the coordinated expression hypothesis for the origin and maintenance of the GAL cluster in yeast
Abstract
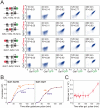
Metabolic gene clusters--functionally related and physically clustered genes--are a common feature of some eukaryotic genomes. Two hypotheses have been advanced to explain the origin and maintenance of metabolic gene clusters: coordinated gene expression and genetic linkage. Here we test the hypothesis that selection for coordinated gene expression underlies the clustering of GAL genes in the yeast genome. We find that, although clustering coordinates the expression of GAL1 and GAL10, disrupting the GAL cluster does not impair fitness, suggesting that other mechanisms, such as genetic linkage, drive the origin and maintenance metabolic gene clusters.
Conflict of interest statement
Figures


References
-
- Field B, Osbourn AE. Metabolic diversification--independent assembly of operon-like gene clusters in different plants. Science. 2008;320:543–547. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

