Accounting for diffusion in agent based models of reaction-diffusion systems with application to cytoskeletal diffusion
- PMID: 21966493
- PMCID: PMC3179499
- DOI: 10.1371/journal.pone.0025306
Accounting for diffusion in agent based models of reaction-diffusion systems with application to cytoskeletal diffusion
Abstract
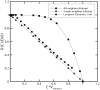
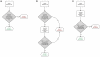
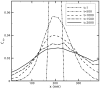
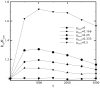
Diffusion plays a key role in many biochemical reaction systems seen in nature. Scenarios where diffusion behavior is critical can be seen in the cell and subcellular compartments where molecular crowding limits the interaction between particles. We investigate the application of a computational method for modeling the diffusion of molecules and macromolecules in three-dimensional solutions using agent based modeling. This method allows for realistic modeling of a system of particles with different properties such as size, diffusion coefficients, and affinity as well as the environment properties such as viscosity and geometry. Simulations using these movement probabilities yield behavior that mimics natural diffusion. Using this modeling framework, we simulate the effects of molecular crowding on effective diffusion and have validated the results of our model using Langevin dynamics simulations and note that they are in good agreement with previous experimental data. Furthermore, we investigate an extension of this framework where single discrete cells can contain multiple particles of varying size in an effort to highlight errors that can arise from discretization that lead to the unnatural behavior of particles undergoing diffusion. Subsequently, we explore various algorithms that differ in how they handle the movement of multiple particles per cell and suggest an algorithm that properly accommodates multiple particles of various sizes per cell that can replicate the natural behavior of these particles diffusing. Finally, we use the present modeling framework to investigate the effect of structural geometry on the directionality of diffusion in the cell cytoskeleton with the observation that parallel orientation in the structural geometry of actin filaments of filopodia and the branched structure of lamellipodia can give directionality to diffusion at the filopodia-lamellipodia interface.
Conflict of interest statement
Figures





References
-
- Dobrzynski M, Rodriguez JV, Kaandorp JA, Blom JG. Computational methods for diffusion-influenced biochemical reactions. Bioinformatics. 2007;23:1969–1977. - PubMed
-
- Gillespie DT. Stochastic simulation of chemical kinetics. Annual Review of Physical Chemistry. 2007;58:35–55. - PubMed
-
- Ridgway D, Broderick G, Ellison MJ. Accommodating space, time and randomness in network simulation. Curr Opin Biotechnol. 2006;17:493–498. - PubMed
-
- Takahashi K, Arjunan SN, Tomita M. Space in systems biology of signaling pathways--towards intracellular molecular crowding in silico. FEBS Lett. 2005;579:1783–1788. - PubMed

