Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts
- PMID: 21966903
- PMCID: PMC2936111
- DOI: 10.1111/j.1462-2920.2009.02065.x
Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts
Abstract
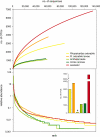
Marine sponges contain complex bacterial communities of considerable ecological and biotechnological importance, with many of these organisms postulated to be specific to sponge hosts. Testing this hypothesis in light of the recent discovery of the rare microbial biosphere, we investigated three Australian sponges by massively parallel 16S rRNA gene tag pyrosequencing. Here we show bacterial diversity that is unparalleled in an invertebrate host, with more than 250,000 sponge-derived sequence tags being assigned to 23 bacterial phyla and revealing up to 2996 operational taxonomic units (95% sequence similarity) per sponge species. Of the 33 previously described 'sponge-specific' clusters that were detected in this study, 48% were found exclusively in adults and larvae - implying vertical transmission of these groups. The remaining taxa, including 'Poribacteria', were also found at very low abundance among the 135,000 tags retrieved from surrounding seawater. Thus, members of the rare seawater biosphere may serve as seed organisms for widely occurring symbiont populations in sponges and their host association might have evolved much more recently than previously thought.
© 2009 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures





References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bayer K, Schmitt S, Hentschel U. Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba. Environ Microbiol. 2008;10:2942–2955. - PubMed
-
- Bock E, Wagner M. Oxidation of inorganic nitrogen compounds as an energy source. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes, Volume 2: Ecophysiology and Biochemistry. New York, USA: Springer Verlag; 2007. pp. 457–495.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

