A role for SUMO in nucleotide excision repair
- PMID: 21968059
- PMCID: PMC3220943
- DOI: 10.1016/j.dnarep.2011.09.013
A role for SUMO in nucleotide excision repair
Abstract
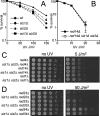
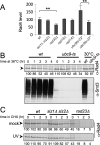
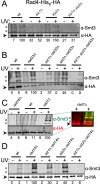
The two Siz/PIAS SUMO E3 ligases Siz1 and Siz2 are responsible for the vast majority of sumoylation in Saccharomyces cerevisiae. We found that siz1Δ siz2Δ mutants are sensitive to ultra-violet (UV) light. Epistasis analysis showed that the SIZ genes act in the nucleotide excision repair (NER) pathway, and suggested that they participate both in global genome repair (GGR) and in the Rpb9-dependent subpathway of transcription-coupled repair (TCR), but have minimal role in Rad26-dependent TCR. Quantitative analysis of NER at the single-nucleotide level showed that siz1Δ siz2Δ is deficient in repair of both the transcribed and non-transcribed strands of the DNA. These experiments confirmed that the SIZ genes participate in GGR. Their role in TCR remains unclear. It has been reported previously that mutants deficient for the SUMO conjugating enzyme Ubc9 contain reduced levels of Rad4, the yeast homolog of human XPC. However, our experiments do not support the conclusion that SUMO conjugation affects Rad4 levels. We found that several factors that participate in NER are sumoylated, including Rad4, Rad16, Rad7, Rad1, Rad10, Ssl2, Rad3, and Rpb4. Although Rad16 was heavily sumoylated, elimination of the major SUMO attachment sites in Rad16 had no detectable effect on UV resistance or removal of DNA lesions. SUMO attachment to most of these NER factors was significantly increased by DNA damage. Furthermore, SUMO-modified Rad4 accumulated in NER mutants that block the pathway downstream of Rad4, suggesting that SUMO becomes attached to Rad4 at a specific point during its functional cycle. Collectively, these results suggest that SIZ-dependent sumoylation may modulate the activity of multiple proteins to promote efficient NER.
2011 Elsevier B.V. All rights reserved.
Figures




References
-
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. - PubMed
-
- Thomson TM, Guerra-Rebollo M. Ubiquitin and SUMO signalling in DNA repair. Biochem Soc Trans. 2010;38:116–131. - PubMed
-
- Ulrich HD. The SUMO system: an overview. Methods Mol Biol. 2009;497:3–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

