The potter's wheel: the host's role in sculpting its microbiota
- PMID: 21968920
- PMCID: PMC3222938
- DOI: 10.1007/s00018-011-0830-3
The potter's wheel: the host's role in sculpting its microbiota
Abstract
Animals, ranging from basal metazoans to primates, are host to complex microbial ecosystems; engaged in a symbiotic relationship that is essential for host physiology and homeostasis. Epithelial surfaces vary in the composition of colonizing microbiota as one compares anatomic sites, developmental stages and species origin. Alterations of microbial composition likely contribute to susceptibility to several distinct diseases. The forces that shape the colonizing microbial composition are the focus of much current investigation, and it is evident that there are pressures exerted both by the host and the external environment to mold these ecosystems. The focus of this review is to discuss recent studies that demonstrate the critical importance of host factors in selecting for its microbiome. Greater insight into host-microbiome interactions will be essential for understanding homeostasis at mucosal surfaces, and developing useful interventions when homeostasis is disrupted.
Figures

 ); Bacteroidetes, green (
); Bacteroidetes, green ( ); Fusobacteria, purple (
); Fusobacteria, purple ( ); Proteobacteria, black (
); Proteobacteria, black ( ); Cyanobacteria, light green (
); Cyanobacteria, light green ( ); Actinobacteria, red (
); Actinobacteria, red ( )
)
 ); β-Proteobacteria, light blue (
); β-Proteobacteria, light blue ( ); δ-Proteobacteria, white (
); δ-Proteobacteria, white ( ); γ-Proteobacteria, black (
); γ-Proteobacteria, black ( ); Spirochetes, pink (
); Spirochetes, pink ( ); Bacteroidetes, green (
); Bacteroidetes, green ( )
)
 ); Firmicutes, yellow (
); Firmicutes, yellow ( ); Bacteroidetes, green (
); Bacteroidetes, green ( ); Fusobacteria, purple (
); Fusobacteria, purple ( ); Actinobacteria, red (
); Actinobacteria, red ( ); Planctomycetes, tan (
); Planctomycetes, tan ( ). The tree was based on pairwise differences (weighted UniFrac metric) between the four communities
). The tree was based on pairwise differences (weighted UniFrac metric) between the four communities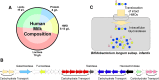



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

