Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events
- PMID: 21969370
- PMCID: PMC3308894
- DOI: 10.1074/jbc.M111.301465
Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events
Abstract
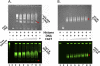
In eukaryotic cells, DNA maintenance requires ordered disassembly and re-assembly of chromatin templates. These processes are highly regulated and require extrinsic factors such as chromatin remodelers and histone chaperones. The histone chaperone FACT (facilitates chromatin transcription) is a large heterodimeric complex with roles in transcription, replication, and repair. FACT promotes and subsequently restricts access to DNA as a result of dynamic nucleosome reorganization. However, until now, there lacked a truly quantitative assessment of the critical contacts mediating FACT function. Here, we demonstrate that FACT binds histones, DNA, and intact nucleosomes at nanomolar concentrations. We also determine roles for the histone tails in free histone and nucleosome binding by FACT. Furthermore, we propose that the conserved acidic C-terminal domain of the FACT subunit Spt16 actively displaces nucleosomal DNA to provide access to the histone octamer. Experiments with tri-nucleosome arrays indicate a possible mode for FACT binding within chromatin. Together, the data reveal that specific FACT subunits synchronize interactions with various target sites on individual nucleosomes to generate a high affinity binding event and promote reorganization.
Figures
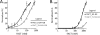
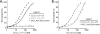
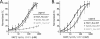


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

