A metagenomic study of methanotrophic microorganisms in Coal Oil Point seep sediments
- PMID: 21970369
- PMCID: PMC3197505
- DOI: 10.1186/1471-2180-11-221
A metagenomic study of methanotrophic microorganisms in Coal Oil Point seep sediments
Abstract
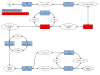
Background: Methane oxidizing prokaryotes in marine sediments are believed to function as a methane filter reducing the oceanic contribution to the global methane emission. In the anoxic parts of the sediments, oxidation of methane is accomplished by anaerobic methanotrophic archaea (ANME) living in syntrophy with sulphate reducing bacteria. This anaerobic oxidation of methane is assumed to be a coupling of reversed methanogenesis and dissimilatory sulphate reduction. Where oxygen is available aerobic methanotrophs take part in methane oxidation. In this study, we used metagenomics to characterize the taxonomic and metabolic potential for methane oxidation at the Tonya seep in the Coal Oil Point area, California. Two metagenomes from different sediment depth horizons (0-4 cm and 10-15 cm below sea floor) were sequenced by 454 technology. The metagenomes were analysed to characterize the distribution of aerobic and anaerobic methanotrophic taxa at the two sediment depths. To gain insight into the metabolic potential the metagenomes were searched for marker genes associated with methane oxidation.
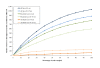
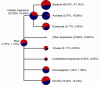
Results: Blast searches followed by taxonomic binning in MEGAN revealed aerobic methanotrophs of the genus Methylococcus to be overrepresented in the 0-4 cm metagenome compared to the 10-15 cm metagenome. In the 10-15 cm metagenome, ANME of the ANME-1 clade, were identified as the most abundant methanotrophic taxon with 8.6% of the reads. Searches for particulate methane monooxygenase (pmoA) and methyl-coenzyme M reductase (mcrA), marker genes for aerobic and anaerobic oxidation of methane respectively, identified pmoA in the 0-4 cm metagenome as Methylococcaceae related. The mcrA reads from the 10-15 cm horizon were all classified as originating from the ANME-1 clade.
Conclusions: Most of the taxa detected were present in both metagenomes and differences in community structure and corresponding metabolic potential between the two samples were mainly due to abundance differences. The results suggests that the Tonya Seep sediment is a robust methane filter, where taxa presently dominating this process could be replaced by less abundant methanotrophic taxa in case of changed environmental conditions.
Figures



References
-
- Hornafius JS, Quigley D, Luyendyk BP. The world's most spectacular marine hydrocarbon seeps (Coal Oil Point, Santa Barbara Channel, California): Quantification of emissions. J Geophys Res. 1999;104(C9):20703–20711. doi: 10.1029/1999JC900148. - DOI
-
- Boles JR, Eichhubl P, Garven G, Chen J. Evolution of a hydrocarbon migration pathway along basin-bounding faults: Evidence from fault cement. Am Assoc Pet Geol Bull. 2004;88(7):947–970.
-
- Luyendyk B, Kennett J, Clark JF. Hypothesis for increased atmospheric methane input from hydrocarbon seeps on exposed continental shelves during glacial low sea level. Marine and Petroleum Geology. 2005;22(4):591–596. doi: 10.1016/j.marpetgeo.2004.08.005. - DOI
-
- Reeburgh WS. ''Soft spots'' in the global methane budget. Microbial Growth on C1 Compounds. 1996. pp. 334–342.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

