Genome-wide association study of coronary artery disease in the Japanese
- PMID: 21971053
- PMCID: PMC3283177
- DOI: 10.1038/ejhg.2011.184
Genome-wide association study of coronary artery disease in the Japanese
Abstract
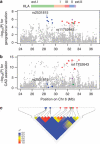
A new understanding of the genetic basis of coronary artery disease (CAD) has recently emerged from genome-wide association (GWA) studies of common single-nucleotide polymorphisms (SNPs), thus far performed mostly in European-descent populations. To identify novel susceptibility gene variants for CAD and confirm those previously identified mostly in populations of European descent, a multistage GWA study was performed in the Japanese. In the discovery phase, we first genotyped 806 cases and 1337 controls with 451 382 SNP markers and subsequently assessed 34 selected SNPs with direct genotyping (541 additional cases) and in silico comparison (964 healthy controls). In the replication phase, involving 3052 cases and 6335 controls, 12 SNPs were tested; CAD association was replicated and/or verified for 4 (of 12) SNPs from 3 loci: near BRAP and ALDH2 on 12q24 (P=1.6 × 10(-34)), HLA-DQB1 on 6p21 (P=4.7 × 10(-7)), and CDKN2A/B on 9p21 (P=6.1 × 10(-16)). On 12q24, we identified the strongest association signal with the strength of association substantially pronounced for a subgroup of myocardial infarction cases (P=1.4 × 10(-40)). On 6p21, an HLA allele, DQB1(*)0604, could show one of the most prominent association signals in an ∼8-Mb interval that encompasses the LTA gene, where an association with myocardial infarction had been reported in another Japanese study. CAD association was also identified at CDKN2A/B, as previously reported in different populations of European descent and Asians. Thus, three loci confirmed in the Japanese GWA study highlight the likely presence of risk alleles with two types of genetic effects - population specific and common - on susceptibility to CAD.
Figures



References
-
- Schunkert H, Erdmann J, Samani NJ. Genetics of myocardial infarction: a progress report. Eur Heart J. 2010;31:918–925. - PubMed
-
- Trégouët DA, König IR, Erdmann J, et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

