Fundamental reaction pathway and free energy profile for hydrolysis of intracellular second messenger adenosine 3',5'-cyclic monophosphate (cAMP) catalyzed by phosphodiesterase-4
- PMID: 21973014
- PMCID: PMC3209513
- DOI: 10.1021/jp205509w
Fundamental reaction pathway and free energy profile for hydrolysis of intracellular second messenger adenosine 3',5'-cyclic monophosphate (cAMP) catalyzed by phosphodiesterase-4
Abstract
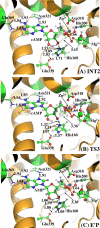
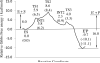
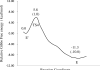
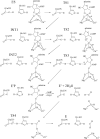
As important drug targets for a variety of human diseases, cyclic nucleotide phosphodiesterases (PDEs) are a superfamily of enzymes sharing a similar catalytic site. We have performed pseudobond first-principles quantum mechanical/molecular mechanical-free energy perturbation (QM/MM-FE) and QM/MM-Poisson-Boltzmann surface area (PBSA) calculations to uncover the detailed reaction mechanism for PDE4-catalyzed hydrolysis of adenosine 3',5'-cyclic monophosphate (cAMP). This is the first report on QM/MM reaction-coordinate calculations including the protein environment of any PDE-catalyzed reaction system, demonstrating a unique catalytic reaction mechanism. The QM/MM-FE and QM/MM-PBSA calculations revealed that the PDE4-catalyzed hydrolysis of cAMP consists of two reaction stages: cAMP hydrolysis (stage 1) and bridging hydroxide ion regeneration (stage 2). The stage 1 includes the binding of cAMP in the active site, nucleophilic attack of the bridging hydroxide ion on the phosphorus atom of cAMP, cleavage of O3'-P phosphoesteric bond of cAMP, protonation of the departing O3' atom, and dissociation of hydrolysis product (AMP). The stage 2 includes the binding of solvent water molecules with the metal ions in the active site and regeneration of the bridging hydroxide ion. The dissociation of the hydrolysis product is found to be rate-determining for the enzymatic reaction process. The calculated activation Gibbs free energy of ≥16.0 and reaction free energy of -11.1 kcal/mol are in good agreement with the experimentally derived activation free energy of 16.6 kcal/mol and reaction free energy of -11.5 kcal/mol, suggesting that the catalytic mechanism obtained from this study is reliable and provides a solid base for future rational drug design.
Figures


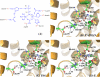
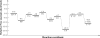
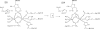
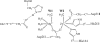
References
-
- Callahan SM, Cornell NW, Dunlap PV. J. Biol. Chem. 1995;270(29):17627–17632. - PubMed
-
- Xu RX, Hassell AM, Vanderwall D, Lambert MH, Holmes WD, Luther MA, Rocque WJ, Milburn MV, Zhao YD, Ke HM, Nolte RT. Science. 2000;288(5472):1822–1825. - PubMed
-
- Ke HM, Wang HC. Curr. Top. Med. Chem. 2007;7(4):391–403. - PubMed
-
- Conti M, Jin SLC, Monaco L, Repaske DR, Swinnen JV. Endocr. Rev. 1991;12(3):218–234. - PubMed
-
- Teixeira MM, Gristwood RW, Cooper N, Hellewell PG. Trend. Pharmacol. Sci. 1997;18(5):164–170. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

