Evolution of vacuolar proton pyrophosphatase domains and volutin granules: clues into the early evolutionary origin of the acidocalcisome
- PMID: 21974828
- PMCID: PMC3198990
- DOI: 10.1186/1745-6150-6-50
Evolution of vacuolar proton pyrophosphatase domains and volutin granules: clues into the early evolutionary origin of the acidocalcisome
Abstract
Background: Volutin granules appear to be universally distributed and are morphologically and chemically identical to acidocalcisomes, which are electron-dense granular organelles rich in calcium and phosphate, whose functions include storage of phosphorus and various metal ions, metabolism of polyphosphate, maintenance of intracellular pH, osmoregulation and calcium homeostasis. Prokaryotes are thought to differ from eukaryotes in that they lack membrane-bounded organelles. However, it has been demonstrated that as in acidocalcisomes, the calcium and polyphosphate-rich intracellular "volutin granules (polyphosphate bodies)" in two bacterial species, Agrobacterium tumefaciens, and Rhodospirillum rubrum, are membrane bound and that the vacuolar proton-translocating pyrophosphatases (V-H+PPases) are present in their surrounding membranes. Volutin granules and acidocalcisomes have been found in organisms as diverse as bacteria and humans.
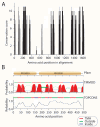
Results: Here, we show volutin granules also occur in Archaea and are, therefore, present in the three superkingdoms of life (Archaea, Bacteria and Eukarya). Molecular analyses of V-H+PPase pumps, which acidify the acidocalcisome lumen and are diagnostic proteins of the organelle, also reveal the presence of this enzyme in all three superkingdoms suggesting it is ancient and universal. Since V-H+PPase sequences contained limited phylogenetic signal to fully resolve the ancestral nodes of the tree, we investigated the divergence of protein domains in the V-H+PPase molecules. Using Protein family (Pfam) database, we found a domain in the protein, PF03030. The domain is shared by 31 species in Eukarya, 231 in Bacteria, and 17 in Archaea. The universal distribution of the V-H+PPase PF03030 domain, which is associated with the V-H+PPase function, suggests the domain and the enzyme were already present in the Last Universal Common Ancestor (LUCA).
Conclusion: The importance of the V-H+PPase function and the evolutionary dynamics of these domains support the early origin of the acidocalcisome organelle. In particular, the universality of volutin granules and presence of a functional V-H+PPase domain in the three superkingdoms of life reveals that the acidocalcisomes may have appeared earlier than the divergence of the superkingdoms. This result is remarkable and highlights the possibility that a high degree of cellular compartmentalization could already have been present in the LUCA.
Figures

References
-
- Margulis L. Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp Soc Exp Biol. 1975. pp. 21–38. - PubMed
-
- Martin W, Russell MJ. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci. 2003;358:59–83. doi: 10.1098/rstb.2002.1183. discussion 83-55. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

