Vector autoregression, structural equation modeling, and their synthesis in neuroimaging data analysis
- PMID: 21975109
- PMCID: PMC3223325
- DOI: 10.1016/j.compbiomed.2011.09.004
Vector autoregression, structural equation modeling, and their synthesis in neuroimaging data analysis
Abstract
Vector autoregression (VAR) and structural equation modeling (SEM) are two popular brain-network modeling tools. VAR, which is a data-driven approach, assumes that connected regions exert time-lagged influences on one another. In contrast, the hypothesis-driven SEM is used to validate an existing connectivity model where connected regions have contemporaneous interactions among them. We present the two models in detail and discuss their applicability to FMRI data, and their interpretational limits. We also propose a unified approach that models both lagged and contemporaneous effects. The unifying model, structural vector autoregression (SVAR), may improve statistical and explanatory power, and avoid some prevalent pitfalls that can occur when VAR and SEM are utilized separately.
Published by Elsevier Ltd.
Figures



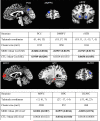
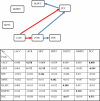


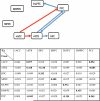


References
-
- Gitelman DR, Penny WD, Ashburner J, Friston KJ. Modeling regional and psychophysiologic interactions in fMRI: the importance of hemodynamic deconvolution. NeuroImage. 2003;19:200–207. - PubMed
-
- Roebroeck A, Formisano E, Goebel R. Mapping directed influence over the brain using granger causality mapping. NeuroImage. 2005;25:230–242. - PubMed
-
- McIntosh AR, Gonzalez-Lima F. Structural equation modeling and its application to network analysis in functional brain imaging. Hum. Brain Mapp. 1994;2:2–22.
-
- Friston KJ, Harrison L, Penny W. Dynamic causal modeling. Neuroimage. 2003;19:1273–1302. - PubMed
-
- Friston KJ. Functional integration. In: Frackowiak RSJ, Friston KJ, Frith C, Dolan R, Friston KJ, Price CJ, Zeki S, Ashburner J, Penny WD, editors. Human Brain Function. 2nd ed. Academic Press; 2003.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

