E4BP4 facilitates glucocorticoid-evoked apoptosis of human leukemic CEM cells via upregulation of Bim
- PMID: 21975218
- PMCID: PMC3197565
- DOI: 10.1186/1750-2187-6-13
E4BP4 facilitates glucocorticoid-evoked apoptosis of human leukemic CEM cells via upregulation of Bim
Abstract
Background: Synthetic GCs serve as therapeutic agents for some lymphoid leukemias because of their ability to induce transcriptional changes via the GC receptor (GR) and trigger apoptosis. Upregulation of the BH3-only member of Bcl-2 family proteins, Bim, has been shown to be essential for GC-evoked apoptosis of leukemic lymphoblasts. Using human T cell leukemic sister clones CEM-C7-14 and CEM-C1-15, we have previously shown that the bZIP transcriptional repressor, E4BP4, is preferentially upregulated by GCs in CEM-C7-14 cells that are susceptible to GC-evoked apoptosis, but not in refractory CEM-C1-15 cells. E4BP4 is an evolutionarily conserved member of the PAR family of bZIP transcription factors related to the C. elegans death specification gene ces2.
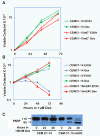
Results: Mouse E4BP4 was ectopically expressed in CEM-C1-15 cells, resulting in sensitization to GC-evoked apoptosis in correlation with restoration of E4BP4 and Bim upregulation. shRNA mediated modest knockdown of E4BP4 in CEM-C7-14 cells resulted in concomitant reduction in Bim expression, although GC-evoked fold-induction and sensitivity to apoptosis was similar to parental cells.
Conclusion: Data presented here suggest that GC-mediated upregulation of E4BP4 facilitates Bim upregulation and apoptosis of CEM cells. Since the Bim promoter does not contain any consensus GRE or EBPRE sequences, induction of Bim may be a secondary response.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

