Cellular poly(c) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1β and support viral replication
- PMID: 21976648
- PMCID: PMC3233143
- DOI: 10.1128/JVI.05177-11
Cellular poly(c) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1β and support viral replication
Abstract
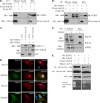
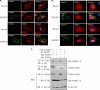
Porcine reproductive and respiratory syndrome virus (PRRSV) infection of swine results in substantial economic losses to the swine industry worldwide. Identification of cellular factors involved in PRRSV life cycle not only will enable a better understanding of virus biology but also has the potential for the development of antiviral therapeutics. The PRRSV nonstructural protein 1 (nsp1) has been shown to be involved in at least two important functions in the infected hosts: (i) mediation of viral subgenomic (sg) mRNA transcription and (ii) suppression of the host's innate immune response mechanisms. To further our understanding of the role of the viral nsp1 in these processes, using nsp1β, a proteolytically processed functional product of nsp1 as bait, we have identified the cellular poly(C)-binding proteins 1 and 2 (PCBP1 and PCBP2) as two of its interaction partners. The interactions of PCBP1 and PCBP2 with nsp1β were confirmed both by coimmunoprecipitation in infected cells and/or in plasmid-transfected cells and also by in vitro binding assays. During PRRSV infection of MARC-145 cells, the cytoplasmic PCBP1 and PCBP2 partially colocalize to the viral replication-transcription complexes. Furthermore, recombinant purified PCBP1 and PCBP2 were found to bind the viral 5' untranslated region (5'UTR). Small interfering RNA (siRNA)-mediated silencing of PCBP1 and PCBP2 in cells resulted in significantly reduced PRRSV genome replication and transcription without adverse effect on initial polyprotein synthesis. Overall, the results presented here point toward an important role for PCBP1 and PCBP2 in regulating PRRSV RNA synthesis.
Figures

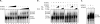



Similar articles
-
Interaction of cellular poly(C)-binding protein 2 with nonstructural protein 1β is beneficial to Chinese highly pathogenic porcine reproductive and respiratory syndrome virus replication.Virus Res. 2012 Oct;169(1):222-30. doi: 10.1016/j.virusres.2012.08.002. Epub 2012 Aug 20. Virus Res. 2012. PMID: 22951310
-
Molecular characterization of the RNA-protein complex directing -2/-1 programmed ribosomal frameshifting during arterivirus replicase expression.J Biol Chem. 2020 Dec 25;295(52):17904-17921. doi: 10.1074/jbc.RA120.016105. Epub 2020 Oct 30. J Biol Chem. 2020. PMID: 33127640 Free PMC article.
-
Interleukin-2 enhancer binding factor 2 interacts with the nsp9 or nsp2 of porcine reproductive and respiratory syndrome virus and exerts negatively regulatory effect on the viral replication.Virol J. 2017 Jul 11;14(1):125. doi: 10.1186/s12985-017-0794-5. Virol J. 2017. PMID: 28693575 Free PMC article.
-
The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins.Virus Res. 2010 Dec;154(1-2):61-76. doi: 10.1016/j.virusres.2010.07.030. Epub 2010 Aug 7. Virus Res. 2010. PMID: 20696193 Free PMC article. Review.
-
Modulation of innate immune signaling by nonstructural protein 1 (nsp1) in the family Arteriviridae.Virus Res. 2014 Dec 19;194:100-9. doi: 10.1016/j.virusres.2014.09.007. Epub 2014 Sep 28. Virus Res. 2014. PMID: 25262851 Free PMC article. Review.
Cited by
-
Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use.Nucleic Acids Res. 2016 Sep 6;44(15):7007-78. doi: 10.1093/nar/gkw530. Epub 2016 Jul 19. Nucleic Acids Res. 2016. PMID: 27436286 Free PMC article. Review.
-
Cellular poly(C) binding protein 2 interacts with porcine epidemic diarrhea virus papain-like protease 1 and supports viral replication.Vet Microbiol. 2020 Aug;247:108793. doi: 10.1016/j.vetmic.2020.108793. Epub 2020 Jul 13. Vet Microbiol. 2020. PMID: 32768236 Free PMC article.
-
Deciphering Immune Modulation in Chickens Co-Infected with ALV-J and CIAV: A Transcriptomic Approach.Microorganisms. 2024 Nov 28;12(12):2453. doi: 10.3390/microorganisms12122453. Microorganisms. 2024. PMID: 39770656 Free PMC article.
-
Transcriptional and Translational Landscape of Equine Torovirus.J Virol. 2018 Aug 16;92(17):e00589-18. doi: 10.1128/JVI.00589-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29950409 Free PMC article.
-
The Identification of Host Proteins That Interact with Non-Structural Proteins-1α and -1β of Porcine Reproductive and Respiratory Syndrome Virus-1.Viruses. 2023 Dec 16;15(12):2445. doi: 10.3390/v15122445. Viruses. 2023. PMID: 38140685 Free PMC article.
References
-
- Andino R., Rieckhof G. E., Baltimore D. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369–380 - PubMed
-
- Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629–633 - PubMed
-
- Celis J. E., et al. (ed.) 2006. Protein detection in gels by silver staining: a procedure compatible with mass-spectrometry, 3rd ed., vol. 4 Elsevier, New York, NY
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

