Infection with seasonal influenza virus elicits CD4 T cells specific for genetically conserved epitopes that can be rapidly mobilized for protective immunity to pandemic H1N1 influenza virus
- PMID: 21976658
- PMCID: PMC3233140
- DOI: 10.1128/JVI.05728-11
Infection with seasonal influenza virus elicits CD4 T cells specific for genetically conserved epitopes that can be rapidly mobilized for protective immunity to pandemic H1N1 influenza virus
Abstract
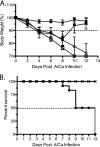
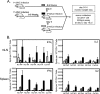
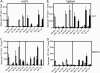
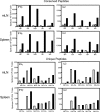
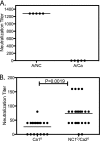
In recent years, influenza viruses with pandemic potential have been a major concern worldwide. One unresolved issue is how infection or vaccination with seasonal influenza virus strains influences the ability to mount a protective immune response to novel pandemic strains. In this study, we developed a mouse model of primary and secondary influenza infection by using a widely circulating seasonal H1N1 virus and the pandemic strain of H1N1 that emerged in Mexico in 2009, and we evaluated several key issues. First, using overlapping peptide libraries encompassing the entire translated sequences of 5 major influenza virus proteins, we assessed the specificity of CD4 T cell reactivity toward epitopes conserved among H1N1 viruses or unique to the seasonal or pandemic strain by enzyme-linked immunospot (ELISpot) assays. Our data show that CD4 T cells reactive to both virus-specific and genetically conserved epitopes are elicited, allowing separate tracking of these responses. Populations of cross-reactive CD4 T cells generated from seasonal influenza infection were found to expand earlier after secondary infection with the pandemic H1N1 virus than CD4 T cell populations specific for new epitopes. Coincident with this rapid CD4 T cell response was a potentiated neutralizing-antibody response to the pandemic strain and protection from the pathological effects of infection with the pandemic virus. This protection was not dependent on CD8 T cells. Together, our results indicate that exposure to seasonal vaccines and infection elicits CD4 T cells that promote the ability of the mammalian host to mount a protective immune response to pandemic strains of influenza virus.
Figures


References
-
- Alberts R., et al. 2010. Gene expression changes in the host response between resistant and susceptible inbred mouse strains after influenza A infection. Microbes Infect. 12:309–318 - PubMed
-
- Albright F. S., Orlando P., Pavia A. T., Jackson G. G., Cannon Albright L. A. 2008. Evidence for a heritable predisposition to death due to influenza. J. Infect. Dis. 197:18–24 - PubMed
-
- Ansaldi F., Durando P., Icardi G. 2011. Intradermal influenza vaccine and new devices: a promising chance for vaccine improvement. Expert Opin. Biol. Ther. 11:415–427 - PubMed
-
- Bautista E., et al. 2010. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. N. Engl. J. Med. 362:1708–1719 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

