Efnb1 and Efnb2 proteins regulate thymocyte development, peripheral T cell differentiation, and antiviral immune responses and are essential for interleukin-6 (IL-6) signaling
- PMID: 21976681
- PMCID: PMC3308828
- DOI: 10.1074/jbc.M111.302596
Efnb1 and Efnb2 proteins regulate thymocyte development, peripheral T cell differentiation, and antiviral immune responses and are essential for interleukin-6 (IL-6) signaling
Abstract
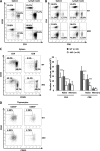
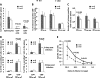
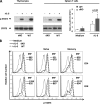
Erythropoietin-producing hepatocellular kinases (Eph kinases) constitute the largest family of cell membrane receptor tyrosine kinases, and their ligand ephrins are also cell surface molecules. Because of promiscuous interaction between Ephs and ephrins, there is considerable redundancy in this system, reflecting the essential roles of these molecules in the biological system through evolution. In this study, both Efnb1 and Efnb2 were null-mutated in the T cell compartment of mice through loxP-mediated gene deletion. Mice with this double conditional mutation (double KO mice) showed reduced thymus and spleen size and cellularity. There was a significant decrease in the DN4, double positive, and single positive thymocyte subpopulations and mature CD4 and CD8 cells in the periphery. dKO thymocytes and peripheral T cells failed to compete with their WT counterparts in irradiated recipients, and the T cells showed compromised ability of homeostatic expansion. dKO naive T cells were inferior in differentiating into Th1 and Th17 effectors in vitro. The dKO mice showed diminished immune response against LCMV infection. Mechanistic studies revealed that IL-6 signaling in dKO T cells was compromised, in terms of abated induction of STAT3 phosphorylation upon IL-6 stimulation. This defect likely contributed to the observed in vitro and in vivo phenotype in dKO mice. This study revealed novel roles of Efnb1 and Efnb2 in T cell development and function.
Figures






References
-
- Eph Nomenclature Committee (1997) Cell 90, 403–404 - PubMed
-
- Wilkinson D. G. (2000) Int. Rev. Cytol. 196, 177–244 - PubMed
-
- Flanagan J. G., Vanderhaeghen P. (1998) Annu. Rev. Neurosci. 21, 309–345 - PubMed
-
- Wang H. U., Chen Z. F., Anderson D. J. (1998) Cell 93, 741–753 - PubMed
-
- Noren N. K., Pasquale E. B. (2007) Cancer Res. 67, 3994–3997 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

