Tagetitoxin inhibits RNA polymerase through trapping of the trigger loop
- PMID: 21976682
- PMCID: PMC3220573
- DOI: 10.1074/jbc.M111.300889
Tagetitoxin inhibits RNA polymerase through trapping of the trigger loop
Abstract
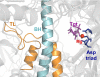
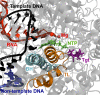
Tagetitoxin (Tgt) inhibits multisubunit chloroplast, bacterial, and some eukaryotic RNA polymerases (RNAPs). A crystallographic structure of Tgt bound to bacterial RNAP apoenzyme shows that Tgt binds near the active site but does not explain why Tgt acts only at certain sites. To understand the Tgt mechanism, we constructed a structural model of Tgt bound to the transcription elongation complex. In this model, Tgt interacts with the β' subunit trigger loop (TL), stabilizing it in an inactive conformation. We show that (i) substitutions of the Arg residue of TL contacted by Tgt confer resistance to inhibitor; (ii) Tgt inhibits RNAP translocation, which requires TL movements; and (iii) paused complexes and a "slow" enzyme, in which the TL likely folds into an altered conformation, are resistant to Tgt. Our studies highlight the role of TL as a target through which accessory proteins and antibiotics can alter the elongation complex dynamics.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

