Non-B DNA Secondary Structures and Their Resolution by RecQ Helicases
- PMID: 21977309
- PMCID: PMC3185257
- DOI: 10.4061/2011/724215
Non-B DNA Secondary Structures and Their Resolution by RecQ Helicases
Abstract
In addition to the canonical B-form structure first described by Watson and Crick, DNA can adopt a number of alternative structures. These non-B-form DNA secondary structures form spontaneously on tracts of repeat sequences that are abundant in genomes. In addition, structured forms of DNA with intrastrand pairing may arise on single-stranded DNA produced transiently during various cellular processes. Such secondary structures have a range of biological functions but also induce genetic instability. Increasing evidence suggests that genomic instabilities induced by non-B DNA secondary structures result in predisposition to diseases. Secondary DNA structures also represent a new class of molecular targets for DNA-interactive compounds that might be useful for targeting telomeres and transcriptional control. The equilibrium between the duplex DNA and formation of multistranded non-B-form structures is partly dependent upon the helicases that unwind (resolve) these alternate DNA structures. With special focus on tetraplex, triplex, and cruciform, this paper summarizes the incidence of non-B DNA structures and their association with genomic instability and emphasizes the roles of RecQ-like DNA helicases in genome maintenance by resolution of DNA secondary structures. In future, RecQ helicases are anticipated to be additional molecular targets for cancer chemotherapeutics.
Figures

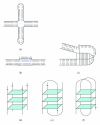

References
-
- Watson JD, Crick FHC. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. - PubMed
-
- Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. Journal of Biological Chemistry. 2004;279(46):47411–47414. - PubMed
-
- Felsenfeld G, Davies DR, Rich A. Formation of a three-stranded polynucleotide molecule. Journal of the American Chemical Society. 1957;79(8):2023–2024.
-
- Wang AHJ, Quigley GJ, Kolpak FJ, et al. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282(5740):680–686. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

