Detergents stabilize the conformation of phosphodiesterase 6
- PMID: 21978030
- PMCID: PMC3215081
- DOI: 10.1021/bi2014695
Detergents stabilize the conformation of phosphodiesterase 6
Abstract
Membrane-bound phosphodiesterase 6 (PDE6) plays an important role in visual signal transduction by regulating cGMP levels in rod photoreceptor cells. Our understanding of PDE6 catalysis and structure suffers from inadequate characterization of the α and β subunit catalytic core, interactions of the core with two intrinsically disordered, proteolysis-prone inhibitory PDEγ (Pγ) subunits, and binding of two types of isoprenyl-binding protein δ, called PrBP/δ, to the isoprenylated C-termini of the catalytic core. Structural studies of native PDE6 have been also been hampered by the lack of a heterologous expression system for the holoenzyme. In this work, we purified PDE6 in the presence of PrBP/δ and screened for additives and detergents that selectively suppress PDE6 basal activity while sparing that of the trypsin-activated enzyme. Some detergents removed PrBP/δ from the PDE complex, separating it from the holoenzyme after PDE6 purification. Additionally, selected detergents also significantly reduced the level of dissociation of PDE6 subunits, increasing their homogeneity and stabilizing the holoenzyme by substituting for its native membrane environment.
Figures
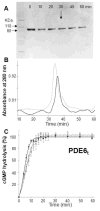


 –) and dimethyldecylphosphine oxide (5 mM, –×–). Those detergents that did not alter PDE6t activity were then tested for their ability to suppress PDE6 activity (lower panel). PDE6 without detergent (solid line), with detergents (dashed lines). N–undecyl–β–D–maltopyranoside (1 mM, –Δ–), C8E4 (10 mM, –○–), Anapoe X–100 (0.8 mM, –◇–). The percentage of cGMP hydrolyzed is plotted vs. time.
–) and dimethyldecylphosphine oxide (5 mM, –×–). Those detergents that did not alter PDE6t activity were then tested for their ability to suppress PDE6 activity (lower panel). PDE6 without detergent (solid line), with detergents (dashed lines). N–undecyl–β–D–maltopyranoside (1 mM, –Δ–), C8E4 (10 mM, –○–), Anapoe X–100 (0.8 mM, –◇–). The percentage of cGMP hydrolyzed is plotted vs. time.



Similar articles
-
Structural characterization of the rod cGMP phosphodiesterase 6.J Mol Biol. 2010 Aug 20;401(3):363-73. doi: 10.1016/j.jmb.2010.06.044. Epub 2010 Jul 1. J Mol Biol. 2010. PMID: 20600113 Free PMC article.
-
Characterization of conformational changes and protein-protein interactions of rod photoreceptor phosphodiesterase (PDE6).J Biol Chem. 2012 Jun 8;287(24):20111-21. doi: 10.1074/jbc.M112.354647. Epub 2012 Apr 18. J Biol Chem. 2012. PMID: 22514270 Free PMC article.
-
Probing the catalytic sites and activation mechanism of photoreceptor phosphodiesterase using radiolabeled phosphodiesterase inhibitors.J Biol Chem. 2009 Nov 13;284(46):31541-7. doi: 10.1074/jbc.M109.018606. Epub 2009 Sep 16. J Biol Chem. 2009. PMID: 19758990 Free PMC article.
-
Photoreceptor phosphodiesterase (PDE6): activation and inactivation mechanisms during visual transduction in rods and cones.Pflugers Arch. 2021 Sep;473(9):1377-1391. doi: 10.1007/s00424-021-02562-x. Epub 2021 Apr 15. Pflugers Arch. 2021. PMID: 33860373 Free PMC article. Review.
-
The retinal cGMP phosphodiesterase gamma-subunit - a chameleon.Curr Protein Pept Sci. 2008 Dec;9(6):611-25. doi: 10.2174/138920308786733930. Curr Protein Pept Sci. 2008. PMID: 19075750 Free PMC article. Review.
Cited by
-
Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases.Sci Adv. 2019 Feb 27;5(2):eaav4322. doi: 10.1126/sciadv.aav4322. eCollection 2019 Feb. Sci Adv. 2019. PMID: 30820458 Free PMC article.
-
A p97/Valosin-Containing Protein Inhibitor Drug CB-5083 Has a Potent but Reversible Off-Target Effect on Phosphodiesterase-6.J Pharmacol Exp Ther. 2021 Jul;378(1):31-41. doi: 10.1124/jpet.120.000486. Epub 2021 Apr 30. J Pharmacol Exp Ther. 2021. PMID: 33931547 Free PMC article.
-
Crystallization of proteins from crude bovine rod outer segments.Methods Enzymol. 2015;557:439-58. doi: 10.1016/bs.mie.2014.11.045. Epub 2015 Mar 17. Methods Enzymol. 2015. PMID: 25950977 Free PMC article.
References
-
- Polans A, Baehr W, Palczewski K. Turned on by Ca2+! The physiology and pathology of Ca(2+)-binding proteins in the retina. Trends Neurosci. 1996;19:547–554. - PubMed
-
- Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. - PubMed
-
- Artemyev NO, Surendran R, Lee JC, Hamm HE. Subunit structure of rod cGMP-phosphodiesterase. J Biol Chem. 1996;271:25382–25388. - PubMed
-
- Baehr W, Devlin MJ, Applebury ML. Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J Biol Chem. 1979;254:11669–11677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

