A proteomic survey of nonribosomal peptide and polyketide biosynthesis in actinobacteria
- PMID: 21978092
- PMCID: PMC3253196
- DOI: 10.1021/pr2009115
A proteomic survey of nonribosomal peptide and polyketide biosynthesis in actinobacteria
Abstract
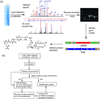
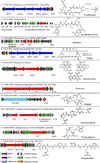
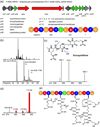
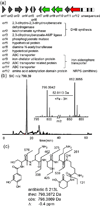
Actinobacteria such as streptomycetes are renowned for their ability to produce bioactive natural products including nonribosomal peptides (NRPs) and polyketides (PKs). The advent of genome sequencing has revealed an even larger genetic repertoire for secondary metabolism with most of the small molecule products of these gene clusters still unknown. Here, we employed a "protein-first" method called PrISM (Proteomic Investigation of Secondary Metabolism) to screen 26 unsequenced actinomycetes using mass spectrometry-based proteomics for the targeted detection of expressed nonribosomal peptide synthetases or polyketide synthases. Improvements to the original PrISM screening approach (Nat. Biotechnol. 2009, 27, 951-956), for example, improved de novo peptide sequencing, have enabled the discovery of 10 NRPS/PKS gene clusters from 6 strains. Taking advantage of the concurrence of biosynthetic enzymes and the secondary metabolites they generate, two natural products were associated with their previously "orphan" gene clusters. This work has demonstrated the feasibility of a proteomics-based strategy for use in screening for NRP/PK production in actinomycetes (often >8 Mbp, high GC genomes) versus the bacilli (2-4 Mbp genomes) used previously.
Figures
References
-
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. - PubMed
-
- Wilkinson B, Micklefield J. Mining and engineering natural-product biosynthetic pathways. Nat Chem Biol. 2007;3:379–386. - PubMed
-
- Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. - PubMed
-
- Van Lanen SG, Shen B. Microbial genomics for the improvement of natural product discovery. Curr Opin Microbiol. 2006;9:252–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

