Conservation of lipid functions in cytochrome bc complexes
- PMID: 21978667
- PMCID: PMC3215850
- DOI: 10.1016/j.jmb.2011.09.023
Conservation of lipid functions in cytochrome bc complexes
Abstract
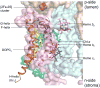
Lipid binding sites and properties are compared in two sub-families of hetero-oligomeric membrane protein complexes known to have similar functions in order to gain further understanding of the role of lipid in the function, dynamics, and assembly of these complexes. Using the crystal structure information for both complexes, we compared the lipid binding properties of the cytochrome b(6)f and bc(1) complexes that function in photosynthetic and respiratory membrane energy transduction. Comparison of lipid and detergent binding sites in the b(6)f complex with those in bc(1) shows significant conservation of lipid positions. Seven lipid binding sites in the cyanobacterial b(6)f complex overlap three natural sites in the Chlamydomonas reinhardtii algal complex and four sites in the yeast mitochondrial bc(1) complex. The specific identity of lipids is different in b(6)f and bc(1) complexes: b(6)f contains sulfoquinovosyldiacylglycerol, phosphatidylglycerol, phosphatidylcholine, monogalactosyldiacylglycerol, and digalactosyldiacylglycerol, whereas cardiolipin, phosphatidylethanolamine, and phosphatidic acid are present in the yeast bc(1) complex. The lipidic chlorophyll a and β-carotene (β-car) in cyanobacterial b(6)f, as well as eicosane in C. reinhardtii, are unique to the b(6)f complex. Inferences of lipid binding sites and functions were supported by sequence, interatomic distance, and B-factor information on interacting lipid groups and coordinating amino acid residues. The lipid functions inferred in the b(6)f complex are as follows: (i) substitution of a transmembrane helix by a lipid and chlorin ring, (ii) lipid and β-car connection of peripheral and core domains, (iii) stabilization of the iron-sulfur protein transmembrane helix, (iv) n-side charge and polarity compensation, and (v) β-car-mediated super-complex with the photosystem I complex.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
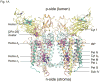
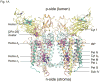
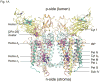
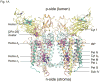







References
-
- Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science (New York, NY. 2003;302:1009–1014. - PubMed
-
- Stroebel D, Choquet Y, Popot JL, Picot D. An atypical heam in the cytochrome b6f complex. Nature. 2003;426:413–418. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

