The NLRP12 pyrin domain: structure, dynamics, and functional insights
- PMID: 21978668
- PMCID: PMC3202057
- DOI: 10.1016/j.jmb.2011.09.024
The NLRP12 pyrin domain: structure, dynamics, and functional insights
Abstract
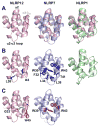
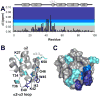
The initial line of defense against infection is sustained by the innate immune system. Together, membrane-bound Toll-like receptors and cytosolic nucleotide-binding domain and leucine-rich repeat-containing receptors (NLR) play key roles in the innate immune response by detecting bacterial and viral invaders as well as endogenous stress signals. NLRs are multi-domain proteins with varying N-terminal effector domains that are responsible for regulating downstream signaling events. Here, we report the structure and dynamics of the N-terminal pyrin domain of NLRP12 (NLRP12 PYD) determined using NMR spectroscopy. NLRP12 is a non-inflammasome NLR that has been implicated in the regulation of Toll-like receptor-dependent nuclear factor-κB activation. NLRP12 PYD adopts a typical six-helical bundle death domain fold. By direct comparison with other PYD structures, we identified hydrophobic residues that are essential for the stable fold of the NLRP PYD family. In addition, we report the first in vitro confirmed non-homotypic PYD interaction between NLRP12 PYD and the pro-apoptotic protein Fas-associated factor 1 (FAF-1), which links the innate immune system to apoptotic signaling. Interestingly, all residues that participate in this protein:protein interaction are confined to the α2-α3 surface, a region of NLRP12 PYD that differs most between currently reported NLRP PYD structures. Finally, we experimentally highlight a significant role for tryptophan 45 in the interaction between NLRP12 PYD and the FAF-1 UBA domain.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
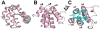




References
-
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–7. - PubMed
-
- Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–59. - PubMed
-
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. - PubMed
-
- Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–54. - PubMed
-
- Faustin B, Reed JC. Sunburned skin activates inflammasomes. Trends Cell Biol. 2008;18:4–8. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

