Multitargeting by curcumin as revealed by molecular interaction studies
- PMID: 21979811
- PMCID: PMC3604998
- DOI: 10.1039/c1np00051a
Multitargeting by curcumin as revealed by molecular interaction studies
Abstract
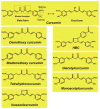
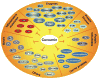
Curcumin (diferuloylmethane), the active ingredient in turmeric (Curcuma longa), is a highly pleiotropic molecule with anti-inflammatory, anti-oxidant, chemopreventive, chemosensitization, and radiosensitization activities. The pleiotropic activities attributed to curcumin come from its complex molecular structure and chemistry, as well as its ability to influence multiple signaling molecules. Curcumin has been shown to bind by multiple forces directly to numerous signaling molecules, such as inflammatory molecules, cell survival proteins, protein kinases, protein reductases, histone acetyltransferase, histone deacetylase, glyoxalase I, xanthine oxidase, proteasome, HIV1 integrase, HIV1 protease, sarco (endo) plasmic reticulum Ca(2+) ATPase, DNA methyltransferases 1, FtsZ protofilaments, carrier proteins, and metal ions. Curcumin can also bind directly to DNA and RNA. Owing to its β-diketone moiety, curcumin undergoes keto-enol tautomerism that has been reported as a favorable state for direct binding. The functional groups on curcumin found suitable for interaction with other macromolecules include the α, β-unsaturated β-diketone moiety, carbonyl and enolic groups of the β-diketone moiety, methoxy and phenolic hydroxyl groups, and the phenyl rings. Various biophysical tools have been used to monitor direct interaction of curcumin with other proteins, including absorption, fluorescence, Fourier transform infrared (FTIR) and circular dichroism (CD) spectroscopy, surface plasmon resonance, competitive ligand binding, Forster type fluorescence resonance energy transfer (FRET), radiolabeling, site-directed mutagenesis, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), immunoprecipitation, phage display biopanning, electron microscopy, 1-anilino-8-naphthalene-sulfonate (ANS) displacement, and co-localization. Molecular docking, the most commonly employed computational tool for calculating binding affinities and predicting binding sites, has also been used to further characterize curcumin's binding sites. Furthermore, the ability of curcumin to bind directly to carrier proteins improves its solubility and bioavailability. In this review, we focus on how curcumin directly targets signaling molecules, as well as the different forces that bind the curcumin-protein complex and how this interaction affects the biological properties of proteins. We will also discuss various analogues of curcumin designed to bind selective targets with increased affinity.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

