Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms
- PMID: 21980304
- PMCID: PMC3183085
- DOI: 10.1371/journal.pgen.1002301
Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms
Abstract
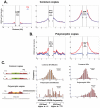
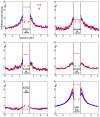
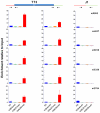
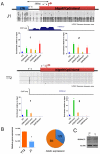
The "arms race" relationship between transposable elements (TEs) and their host has promoted a series of epigenetic silencing mechanisms directed against TEs. Retrotransposons, a class of TEs, are often located in repressed regions and are thought to induce heterochromatin formation and spreading. However, direct evidence for TE-induced local heterochromatin in mammals is surprisingly scarce. To examine this phenomenon, we chose two mouse embryonic stem (ES) cell lines that possess insertionally polymorphic retrotransposons (IAP, ETn/MusD, and LINE elements) at specific loci in one cell line but not the other. Employing ChIP-seq data for these cell lines, we show that IAP elements robustly induce H3K9me3 and H4K20me3 marks in flanking genomic DNA. In contrast, such heterochromatin is not induced by LINE copies and only by a minority of polymorphic ETn/MusD copies. DNA methylation is independent of the presence of IAP copies, since it is present in flanking regions of both full and empty sites. Finally, such spreading into genes appears to be rare, since the transcriptional start sites of very few genes are less than one Kb from an IAP. However, the B3galtl gene is subject to transcriptional silencing via IAP-induced heterochromatin. Hence, although rare, IAP-induced local heterochromatin spreading into nearby genes may influence expression and, in turn, host fitness.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Spreading of heterochromatin is limited to specific families of maize retrotransposons.PLoS Genet. 2012;8(12):e1003127. doi: 10.1371/journal.pgen.1003127. Epub 2012 Dec 13. PLoS Genet. 2012. PMID: 23271981 Free PMC article.
-
Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA.Nat Struct Mol Biol. 2013 Mar;20(3):332-8. doi: 10.1038/nsmb.2495. Epub 2013 Jan 27. Nat Struct Mol Biol. 2013. PMID: 23353788
-
Preferential epigenetic suppression of the autonomous MusD over the nonautonomous ETn mouse retrotransposons.Mol Cell Biol. 2009 May;29(9):2456-68. doi: 10.1128/MCB.01383-08. Epub 2009 Mar 9. Mol Cell Biol. 2009. PMID: 19273603 Free PMC article.
-
Silencing of endogenous retroviruses by heterochromatin.Cell Mol Life Sci. 2017 Jun;74(11):2055-2065. doi: 10.1007/s00018-017-2454-8. Epub 2017 Feb 3. Cell Mol Life Sci. 2017. PMID: 28160052 Free PMC article. Review.
-
Epigenetic fates of gene silencing established by heterochromatin spreading in cell identity and genome stability.Curr Genet. 2019 Apr;65(2):423-428. doi: 10.1007/s00294-018-0901-1. Epub 2018 Nov 2. Curr Genet. 2019. PMID: 30390097 Free PMC article. Review.
Cited by
-
TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells.Genome Res. 2013 Mar;23(3):452-61. doi: 10.1101/gr.147678.112. Epub 2012 Dec 10. Genome Res. 2013. PMID: 23233547 Free PMC article.
-
Transposable element proliferation as a possible side effect of endosymbiont manipulations.Mob Genet Elements. 2012 Sep 1;2(5):253-256. doi: 10.4161/mge.22878. Mob Genet Elements. 2012. PMID: 23550173 Free PMC article.
-
Restricting retrotransposons: a review.Mob DNA. 2016 Aug 11;7:16. doi: 10.1186/s13100-016-0070-z. eCollection 2016. Mob DNA. 2016. PMID: 27525044 Free PMC article. Review.
-
Synergistic epistasis of the deleterious effects of transposable elements.Genetics. 2022 Feb 4;220(2):iyab211. doi: 10.1093/genetics/iyab211. Genetics. 2022. PMID: 34888644 Free PMC article.
-
The Methylation Inhibitor 5-Aza-2'-Deoxycytidine Induces Genome-Wide Hypomethylation in Rice.Rice (N Y). 2022 Jul 2;15(1):35. doi: 10.1186/s12284-022-00580-6. Rice (N Y). 2022. PMID: 35779161 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

