MicroRNA predictors of longevity in Caenorhabditis elegans
- PMID: 21980307
- PMCID: PMC3183074
- DOI: 10.1371/journal.pgen.1002306
MicroRNA predictors of longevity in Caenorhabditis elegans
Abstract
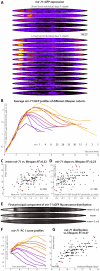
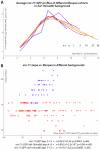
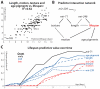
Neither genetic nor environmental factors fully account for variability in individual longevity: genetically identical invertebrates in homogenous environments often experience no less variability in lifespan than outbred human populations. Such variability is often assumed to result from stochasticity in damage accumulation over time; however, the identification of early-life gene expression states that predict future longevity would suggest that lifespan is least in part epigenetically determined. Such "biomarkers of aging," genetic or otherwise, nevertheless remain rare. In this work, we sought early-life differences in organismal robustness in unperturbed individuals and examined the utility of microRNAs, known regulators of lifespan, development, and robustness, as aging biomarkers. We quantitatively examined Caenorhabditis elegans reared individually in a novel apparatus and observed throughout their lives. Early-to-mid-adulthood measures of homeostatic ability jointly predict 62% of longevity variability. Though correlated, markers of growth/muscle maintenance and of metabolic by-products ("age pigments") report independently on lifespan, suggesting that graceful aging is not a single process. We further identified three microRNAs in which early-adulthood expression patterns individually predict up to 47% of lifespan differences. Though expression of each increases throughout this time, mir-71 and mir-246 correlate with lifespan, while mir-239 anti-correlates. Two of these three microRNA "biomarkers of aging" act upstream in insulin/IGF-1-like signaling (IIS) and other known longevity pathways, thus we infer that these microRNAs not only report on but also likely determine longevity. Thus, fluctuations in early-life IIS, due to variation in these microRNAs and from other causes, may determine individual lifespan.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Gögele M, Pattaro C, Fuchsberger C, Minelli C, Pramstaller PP, et al. Heritability Analysis of Life Span in a Semi-isolated Population Followed Across Four Centuries Reveals the Presence of Pleiotropy Between Life Span and Reproduction. J Gerontol A Biol Sci Med Sci. 2010;705:1–12. - PubMed
-
- Herskind AM, McGue M, Holm NV, Sørensen TI, Harvald B, et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. - PubMed
-
- Horiuchi S. Interspecies Differences in the Life Span Distribution: Humans versus Invertebrates. Pop Dev Review. 2003;29:127–151.
-
- Vaupel JW, Carey JR, Christensen K, Johnson TE, Yashin AI, et al. Biodemographic trajectories of longevity. Science. 1998;280:855–860. - PubMed
-
- Brooks A, Lithgow GJ, Johnson TE. Mortality rates in a genetically heterogeneous population of Caenorhabditis elegans. Science. 1994;263:668–671. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

