Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis
- PMID: 21980309
- PMCID: PMC3183091
- DOI: 10.1371/journal.pgen.1002309
Cell type-specific transcriptome analysis reveals a major role for Zeb1 and miR-200b in mouse inner ear morphogenesis
Abstract
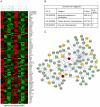
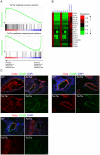
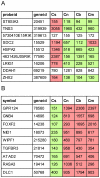
Cellular heterogeneity hinders the extraction of functionally significant results and inference of regulatory networks from wide-scale expression profiles of complex mammalian organs. The mammalian inner ear consists of the auditory and vestibular systems that are each composed of hair cells, supporting cells, neurons, mesenchymal cells, other epithelial cells, and blood vessels. We developed a novel protocol to sort auditory and vestibular tissues of newborn mouse inner ears into their major cellular components. Transcriptome profiling of the sorted cells identified cell type-specific expression clusters. Computational analysis detected transcription factors and microRNAs that play key roles in determining cell identity in the inner ear. Specifically, our analysis revealed the role of the Zeb1/miR-200b pathway in establishing epithelial and mesenchymal identity in the inner ear. Furthermore, we detected a misregulation of the ZEB1 pathway in the inner ear of Twirler mice, which manifest, among other phenotypes, malformations of the auditory and vestibular labyrinth. The association of misregulation of the ZEB1/miR-200b pathway with auditory and vestibular defects in the Twirler mutant mice uncovers a novel mechanism underlying deafness and balance disorders. Our approach can be employed to decipher additional complex regulatory networks underlying other hearing and balance mouse mutants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Friedman TB, Schultz JM, Ben-Yosef T, Pryor SP, Lagziel A, et al. Recent advances in the understanding of syndromic forms of hearing loss. Ear Hear. 2003;24:289–302. - PubMed
-
- Friedman LM, Dror AA, Avraham KB. Mouse models to study inner ear development and hereditary hearing loss. Int J Dev Biol. 2007;51:609–631. - PubMed
-
- Nielsen JS, McNagny KM. Novel functions of the CD34 family. J Cell Sci. 2008;121:3683–3692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

