Antibody V(h) repertoire differences between resolving and chronically evolving hepatitis C virus infections
- PMID: 21980500
- PMCID: PMC3182224
- DOI: 10.1371/journal.pone.0025606
Antibody V(h) repertoire differences between resolving and chronically evolving hepatitis C virus infections
Abstract
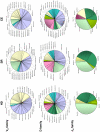
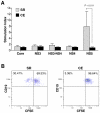
Despite the production of neutralizing antibodies to hepatitis C virus (HCV), many patients fail to clear the virus and instead develop chronic infection and long-term complications. To understand how HCV infection perturbs the antibody repertoire and to identify molecular features of antibody genes associated with either viral clearance or chronic infection, we sequenced the V(D)J region of naïve and memory B cells of 6 persons who spontaneously resolved an HCV infection (SR), 9 patients with a newly diagnosed chronically evolving infection (CE), and 7 healthy donors. In both naïve and memory B cells, the frequency of use of particular antibody gene subfamilies and segments varied among the three clinical groups, especially between SR and CE. Compared to CE, SR antibody genes used fewer VH, D and JH gene segments in naïve B cells and fewer VH segments in memory B cells. SR and CE groups significantly differed in the frequency of use of 7 gene segments in naïve B cell clones and 3 gene segments in memory clones. The nucleotide mutation rates were similar among groups, but the pattern of replacement and silent mutations in memory B cell clones indicated greater antigen selection in SR than CE. Greater clonal evolution of SR than CE memory B cells was revealed by analysis of phylogenetic trees and CDR3 lengths. Pauciclonality of the peripheral memory B cell population is a distinguishing feature of persons who spontaneously resolved an HCV infection. This finding, previously considered characteristic only of patients with HCV-associated lymphoproliferative disorders, suggests that the B cell clones potentially involved in clearance of the virus may also be those susceptible to abnormal expansion.
Conflict of interest statement
Figures




Similar articles
-
Intrahepatic B-cell follicles of chronically hepatitis C virus-infected individuals lack signs of an ectopic germinal center reaction.Eur J Immunol. 2014 Jun;44(6):1842-50. doi: 10.1002/eji.201344378. Epub 2014 Apr 14. Eur J Immunol. 2014. PMID: 24609763 Clinical Trial.
-
Hepatitis C virus (HCV)-induced immunoglobulin hypermutation reduces the affinity and neutralizing activities of antibodies against HCV envelope protein.J Virol. 2008 Jul;82(13):6711-20. doi: 10.1128/JVI.02582-07. Epub 2008 Apr 16. J Virol. 2008. PMID: 18417597 Free PMC article.
-
Hepatitis C virus drives the unconstrained monoclonal expansion of VH1-69-expressing memory B cells in type II cryoglobulinemia: a model of infection-driven lymphomagenesis.J Immunol. 2005 May 15;174(10):6532-9. doi: 10.4049/jimmunol.174.10.6532. J Immunol. 2005. PMID: 15879157
-
Multifaceted functions of B cells in chronic hepatitis C virus infection.Antiviral Res. 2003 Oct;60(2):111-5. doi: 10.1016/j.antiviral.2003.08.004. Antiviral Res. 2003. PMID: 14638406 Review.
-
HCV proteins and immunoglobulin variable gene (IgV) subfamilies in HCV-induced type II mixed cryoglobulinemia: a concurrent pathogenetic role.Clin Dev Immunol. 2012;2012:705013. doi: 10.1155/2012/705013. Epub 2012 May 29. Clin Dev Immunol. 2012. PMID: 22690241 Free PMC article. Review.
Cited by
-
Molecular signature in HCV-positive lymphomas.Clin Dev Immunol. 2012;2012:623465. doi: 10.1155/2012/623465. Epub 2012 Aug 16. Clin Dev Immunol. 2012. PMID: 22952554 Free PMC article. Review.
-
The in Vitro Antigenicity of Plasmodium vivax Rhoptry Neck Protein 2 (PvRON2) B- and T-Epitopes Selected by HLA-DRB1 Binding Profile.Front Cell Infect Microbiol. 2018 May 15;8:156. doi: 10.3389/fcimb.2018.00156. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29868512 Free PMC article.
-
An elaborate landscape of the human antibody repertoire against enterovirus 71 infection is revealed by phage display screening and deep sequencing.MAbs. 2017 Feb/Mar;9(2):342-349. doi: 10.1080/19420862.2016.1267086. Epub 2016 Dec 8. MAbs. 2017. PMID: 27929745 Free PMC article.
-
Early T follicular helper cell activity accelerates hepatitis C virus-specific B cell expansion.J Clin Invest. 2021 Jan 19;131(2):e140590. doi: 10.1172/JCI140590. J Clin Invest. 2021. PMID: 33463551 Free PMC article.
-
The Usage of Human IGHJ Genes Follows a Particular Non-random Selection: The Recombination Signal Sequence May Affect the Usage of Human IGHJ Genes.Front Genet. 2020 Dec 8;11:524413. doi: 10.3389/fgene.2020.524413. eCollection 2020. Front Genet. 2020. PMID: 33363565 Free PMC article.
References
-
- Racanelli V, Rehermann B. Hepatitis C virus infection: when silence is deception. Trends Immunol. 2003;24:456–464. - PubMed
-
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–29. - PubMed
-
- Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–50. - PubMed
-
- Giordano TP, Henderson L, Landgren O, Chiao EY, Kramer JR, et al. Risk of non-Hodgkin lymphoma and lymphoproliferative precursor diseases in US veterans with hepatitis C virus. Jama. 2007;297:2010–2017. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

