UBR1 promotes protein kinase quality control and sensitizes cells to Hsp90 inhibition
- PMID: 21983172
- PMCID: PMC3221935
- DOI: 10.1016/j.yexcr.2011.09.010
UBR1 promotes protein kinase quality control and sensitizes cells to Hsp90 inhibition
Abstract
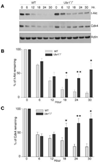
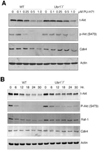
UBR1 and UBR2 are N-recognin ubiquitin ligases that function in the N-end rule degradation pathway. In yeast, the UBR1 homologue also functions by N-end rule independent means to promote degradation of misfolded proteins generated by treatment of cells with geldanamycin, a small molecule inhibitor of Hsp90. Based on these studies we examined the role of mammalian UBR1 and UBR2 in the degradation of protein kinase clients upon Hsp90 inhibition. Our findings show that protein kinase clients Akt and Cdk4 are still degraded in mouse Ubr1(-)/(-) cells treated with geldanamycin, but that their levels recover much more rapidly than is found in wild type cells. These findings correlate with increased induction of Hsp90 expression in the Ubr1(-)/(-) cells compared with wild type cells. We also observed a reduction of UBR1 protein levels in geldanamycin-treated mouse embryonic fibroblasts and human breast cancer cells, suggesting that UBR1 is an Hsp90 client. Further studies revealed a functional overlap between UBR1 and the quality control ubiquitin ligase, CHIP. Our findings show that UBR1 function is conserved in controlling the levels of Hsp90-dependent protein kinases upon geldanamycin treatment, and suggest that it plays a role in determining the sensitivity of cancer cells to the chemotherapeutic effects of Hsp90 inhibitors.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




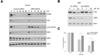

Similar articles
-
A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast.J Biol Chem. 2012 Jul 6;287(28):23911-22. doi: 10.1074/jbc.M112.341164. Epub 2012 May 16. J Biol Chem. 2012. PMID: 22593585 Free PMC article.
-
Molecular mechanism of 17-allylamino-17-demethoxygeldanamycin (17-AAG)-induced AXL receptor tyrosine kinase degradation.J Biol Chem. 2013 Jun 14;288(24):17481-94. doi: 10.1074/jbc.M112.439422. Epub 2013 Apr 29. J Biol Chem. 2013. PMID: 23629654 Free PMC article.
-
Hsp90 rescues PTK6 from proteasomal degradation in breast cancer cells.Biochem J. 2012 Oct 15;447(2):313-20. doi: 10.1042/BJ20120803. Biochem J. 2012. PMID: 22849407
-
Drug-mediated targeted disruption of multiple protein activities through functional inhibition of the Hsp90 chaperone complex.Curr Med Chem. 2007;14(29):3122-38. doi: 10.2174/092986707782793925. Curr Med Chem. 2007. PMID: 18220746 Review.
-
Hsp90 inhibitor geldanamycin and its derivatives as novel cancer chemotherapeutic agents.Curr Pharm Des. 2005;11(9):1131-8. doi: 10.2174/1381612053507585. Curr Pharm Des. 2005. PMID: 15853661 Review.
Cited by
-
The Hsp70 chaperone system: distinct roles in erythrocyte formation and maintenance.Haematologica. 2021 Jun 1;106(6):1519-1534. doi: 10.3324/haematol.2019.233056. Haematologica. 2021. PMID: 33832207 Free PMC article. Review.
-
Bag1 Co-chaperone Promotes TRC8 E3 Ligase-dependent Degradation of Misfolded Human Ether a Go-Go-related Gene (hERG) Potassium Channels.J Biol Chem. 2017 Feb 10;292(6):2287-2300. doi: 10.1074/jbc.M116.752618. Epub 2016 Dec 20. J Biol Chem. 2017. PMID: 27998983 Free PMC article.
-
Specificity in the actions of the UBR1 ubiquitin ligase in the degradation of nuclear receptors.FEBS Open Bio. 2013 Sep 17;3:394-7. doi: 10.1016/j.fob.2013.09.003. eCollection 2013. FEBS Open Bio. 2013. PMID: 24251101 Free PMC article.
-
A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast.J Biol Chem. 2012 Jul 6;287(28):23911-22. doi: 10.1074/jbc.M112.341164. Epub 2012 May 16. J Biol Chem. 2012. PMID: 22593585 Free PMC article.
-
Acquired Glucocorticoid Resistance Due to Homologous Glucocorticoid Receptor Downregulation: A Modern Look at an Age-Old Problem.Cells. 2021 Sep 24;10(10):2529. doi: 10.3390/cells10102529. Cells. 2021. PMID: 34685511 Free PMC article. Review.
References
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
-
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. - PubMed
-
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. - PubMed
-
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

