A primary microcephaly protein complex forms a ring around parental centrioles
- PMID: 21983783
- PMCID: PMC3299569
- DOI: 10.1038/ng.971
A primary microcephaly protein complex forms a ring around parental centrioles
Abstract
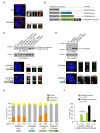
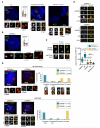
Autosomal recessive primary microcephaly (MCPH) is characterized by a substantial reduction in prenatal human brain growth without alteration of the cerebral architecture and is caused by biallelic mutations in genes coding for a subset of centrosomal proteins. Although at least three of these proteins have been implicated in centrosome duplication, the nature of the centrosome dysfunction that underlies the neurodevelopmental defect in MCPH is unclear. Here we report a homozygous MCPH-causing mutation in human CEP63. CEP63 forms a complex with another MCPH protein, CEP152, a conserved centrosome duplication factor. Together, these two proteins are essential for maintaining normal centrosome numbers in cells. Using super-resolution microscopy, we found that CEP63 and CEP152 co-localize in a discrete ring around the proximal end of the parental centriole, a pattern specifically disrupted in CEP63-deficient cells derived from patients with MCPH. This work suggests that the CEP152-CEP63 ring-like structure ensures normal neurodevelopment and that its impairment particularly affects human cerebral cortex growth.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

