Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3
- PMID: 21983785
- PMCID: PMC3227560
- DOI: 10.1038/ng.958
Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3
Abstract
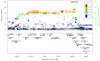
We performed a genome-wide association study of melanoma in a discovery cohort of 2,168 Australian individuals with melanoma and 4,387 control individuals. In this discovery phase, we confirm several previously characterized melanoma-associated loci at MC1R, ASIP and MTAP-CDKN2A. We selected variants at nine loci for replication in three independent case-control studies (Europe: 2,804 subjects with melanoma, 7,618 control subjects; United States 1: 1,804 subjects with melanoma, 1,026 control subjects; United States 2: 585 subjects with melanoma, 6,500 control subjects). The combined meta-analysis of all case-control studies identified a new susceptibility locus at 1q21.3 (rs7412746, P = 9.0 × 10(-11), OR in combined replication cohorts of 0.89 (95% CI 0.85-0.95)). We also show evidence suggesting that melanoma associates with 1q42.12 (rs3219090, P = 9.3 × 10(-8)). The associated variants at the 1q21.3 locus span a region with ten genes, and plausible candidate genes for melanoma susceptibility include ARNT and SETDB1. Variants at the 1q21.3 locus do not seem to be associated with human pigmentation or measures of nevus density.
Figures


References
-
- Gudbjartsson DF, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–91. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- P30CA016672/CA/NCI NIH HHS/United States
- CA87969/CA/NCI NIH HHS/United States
- HG004446/HG/NHGRI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 CA088363/CA/NCI NIH HHS/United States
- U01 CA049449/CA/NCI NIH HHS/United States
- R01 CA133996/CA/NCI NIH HHS/United States
- R01 CA083115/CA/NCI NIH HHS/United States
- CA100264/CA/NCI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- P01 CA055075/CA/NCI NIH HHS/United States
- WT084766/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- R01 CA109544/CA/NCI NIH HHS/United States
- CA49449/CA/NCI NIH HHS/United States
- CA88363/CA/NCI NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- CA83115/CA/NCI NIH HHS/United States
- R01 CA122838/CA/NCI NIH HHS/United States
- CA133996/CA/NCI NIH HHS/United States
- R01CA133996/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 CA049449/CA/NCI NIH HHS/United States
- CA055075/CA/NCI NIH HHS/United States
- C588/A4994/CRUK_/Cancer Research UK/United Kingdom
- P50 CA093459/CA/NCI NIH HHS/United States
- 10589/CRUK_/Cancer Research UK/United Kingdom
- R01 CA100264/CA/NCI NIH HHS/United States
- 2P50CA093459/CA/NCI NIH HHS/United States
- CA109544/CA/NCI NIH HHS/United States
- CA122838/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

