Structure and nucleosome interaction of the yeast NuA4 and Piccolo-NuA4 histone acetyltransferase complexes
- PMID: 21984211
- PMCID: PMC3210417
- DOI: 10.1038/nsmb.2128
Structure and nucleosome interaction of the yeast NuA4 and Piccolo-NuA4 histone acetyltransferase complexes
Abstract
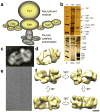
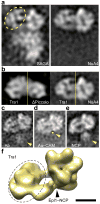
We have used EM and biochemistry to characterize the structure of NuA4, an essential yeast histone acetyltransferase (HAT) complex conserved throughout eukaryotes, and we have determined the interaction of NuA4 with the nucleosome core particle (NCP). The ATM-related Tra1 subunit, which is shared with the SAGA coactivator complex, forms a large domain joined to a second region that accommodates the catalytic subcomplex Piccolo and other NuA4 subunits. EM analysis of a NuA4-NCP complex shows the NCP bound at the periphery of NuA4. EM characterization of Piccolo and Piccolo-NCP provided further information about subunit organization and confirmed that histone acetylation requires minimal contact with the NCP. A small conserved region at the N terminus of Piccolo subunit enhancer of Polycomb-like 1 (Epl1) is essential for NCP interaction, whereas the subunit yeast homolog of mammalian Ing1 2 (Yng2) apparently positions Piccolo for efficient acetylation of histone H4 or histone H2A tails. Taken together, these results provide an understanding of the NuA4 subunit organization and the NuA4-NCP interactions.
Figures

Similar articles
-
Piccolo NuA4-catalyzed acetylation of nucleosomal histones: critical roles of an Esa1 Tudor/chromo barrel loop and an Epl1 enhancer of polycomb A (EPcA) basic region.Mol Cell Biol. 2013 Jan;33(1):159-69. doi: 10.1128/MCB.01131-12. Epub 2012 Oct 29. Mol Cell Biol. 2013. PMID: 23109429 Free PMC article.
-
Site specificity analysis of Piccolo NuA4-mediated acetylation for different histone complexes.Biochem J. 2015 Dec 1;472(2):239-48. doi: 10.1042/BJ20150654. Epub 2015 Sep 29. Biochem J. 2015. PMID: 26420880 Free PMC article.
-
The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes.Mol Cell Biol. 2005 Jul;25(13):5535-42. doi: 10.1128/MCB.25.13.5535-5542.2005. Mol Cell Biol. 2005. PMID: 15964809 Free PMC article.
-
Share and share alike: the role of Tra1 from the SAGA and NuA4 coactivator complexes.Transcription. 2019 Feb;10(1):37-43. doi: 10.1080/21541264.2018.1530936. Epub 2018 Oct 30. Transcription. 2019. PMID: 30375921 Free PMC article. Review.
-
Reading chromatin: insights from yeast into YEATS domain structure and function.Epigenetics. 2010 Oct 1;5(7):573-7. doi: 10.4161/epi.5.7.12856. Epub 2010 Oct 1. Epigenetics. 2010. PMID: 20657183 Free PMC article. Review.
Cited by
-
Histone target selection within chromatin: an exemplary case of teamwork.Genes Dev. 2014 May 15;28(10):1029-41. doi: 10.1101/gad.236331.113. Genes Dev. 2014. PMID: 24831698 Free PMC article. Review.
-
Protein domain mapping by internal labeling and single particle electron microscopy.J Struct Biol. 2015 Nov;192(2):159-62. doi: 10.1016/j.jsb.2015.09.016. Epub 2015 Sep 30. J Struct Biol. 2015. PMID: 26431894 Free PMC article.
-
The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation.Mol Cell. 2016 May 5;62(3):409-421. doi: 10.1016/j.molcel.2016.03.031. Mol Cell. 2016. PMID: 27153538 Free PMC article.
-
A balancing act: interactions within NuA4/TIP60 regulate picNuA4 function in Saccharomyces cerevisiae and humans.Genetics. 2022 Nov 1;222(3):iyac136. doi: 10.1093/genetics/iyac136. Genetics. 2022. PMID: 36066422 Free PMC article.
-
Development of a yeast internal-subunit eGFP labeling strategy and its application in subunit identification in eukaryotic group II chaperonin TRiC/CCT.Sci Rep. 2018 Feb 5;8(1):2374. doi: 10.1038/s41598-017-18962-y. Sci Rep. 2018. PMID: 29403048 Free PMC article.
References
-
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

