Structural basis of cooperativity in human UDP-glucose dehydrogenase
- PMID: 21984906
- PMCID: PMC3184952
- DOI: 10.1371/journal.pone.0025226
Structural basis of cooperativity in human UDP-glucose dehydrogenase
Abstract
Background: UDP-glucose dehydrogenase (UGDH) is the sole enzyme that catalyzes the conversion of UDP-glucose to UDP-glucuronic acid. The product is used in xenobiotic glucuronidation in hepatocytes and in the production of proteoglycans that are involved in promoting normal cellular growth and migration. Overproduction of proteoglycans has been implicated in the progression of certain epithelial cancers, while inhibition of UGDH diminished tumor angiogenesis in vivo. A better understanding of the conformational changes occurring during the UGDH reaction cycle will pave the way for inhibitor design and potential cancer therapeutics.
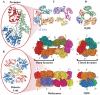
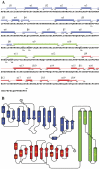
Methodology: Previously, the substrate-bound of UGDH was determined to be a symmetrical hexamer and this regular symmetry is disrupted on binding the inhibitor, UDP-α-D-xylose. Here, we have solved an alternate crystal structure of human UGDH (hUGDH) in complex with UDP-glucose at 2.8 Å resolution. Surprisingly, the quaternary structure of this substrate-bound protein complex consists of the open homohexamer that was previously observed for inhibitor-bound hUGDH, indicating that this conformation is relevant for deciphering elements of the normal reaction cycle.
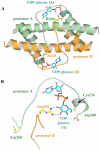
Conclusion: In all subunits of the present open structure, Thr131 has translocated into the active site occupying the volume vacated by the absent active water and partially disordered NAD+ molecule. This conformation suggests a mechanism by which the enzyme may exchange NADH for NAD+ and repolarize the catalytic water bound to Asp280 while protecting the reaction intermediates. The structure also indicates how the subunits may communicate with each other through two reaction state sensors in this highly cooperative enzyme.
Conflict of interest statement
Figures


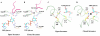
Similar articles
-
Hysteresis and negative cooperativity in human UDP-glucose dehydrogenase.Biochemistry. 2013 Feb 26;52(8):1456-65. doi: 10.1021/bi301593c. Epub 2013 Feb 14. Biochemistry. 2013. PMID: 23363239
-
Cofactor binding triggers a molecular switch to allosterically activate human UDP-α-D-glucose 6-dehydrogenase.Biochemistry. 2012 Nov 20;51(46):9364-74. doi: 10.1021/bi301067w. Epub 2012 Nov 8. Biochemistry. 2012. PMID: 23106432
-
Structure and mechanism of human UDP-glucose 6-dehydrogenase.J Biol Chem. 2011 Jul 8;286(27):23877-87. doi: 10.1074/jbc.M111.234682. Epub 2011 Apr 18. J Biol Chem. 2011. PMID: 21502315 Free PMC article.
-
UDP-glucose dehydrogenase: structure and function of a potential drug target.Biochem Soc Trans. 2010 Oct;38(5):1378-85. doi: 10.1042/BST0381378. Biochem Soc Trans. 2010. PMID: 20863317 Review.
-
Catalytic mechanism of UDP-glucose dehydrogenase.Biochem Soc Trans. 2019 Jun 28;47(3):945-955. doi: 10.1042/BST20190257. Epub 2019 Jun 12. Biochem Soc Trans. 2019. PMID: 31189734 Review.
Cited by
-
UDP-glucose dehydrogenase (UGDH) in clinical oncology and cancer biology.Oncotarget. 2023 Sep 28;14:843-857. doi: 10.18632/oncotarget.28514. Oncotarget. 2023. PMID: 37769033 Free PMC article. Review.
-
In silico prediction of targets for anti-angiogenesis and their in vitro evaluation confirm the involvement of SOD3 in angiogenesis.Oncotarget. 2018 Apr 3;9(25):17349-17367. doi: 10.18632/oncotarget.24693. eCollection 2018 Apr 3. Oncotarget. 2018. PMID: 29707113 Free PMC article.
-
Elevated UDP-glucuronic acid levels mend drug resistance and stress responses via a protease and a transporter in Cryptococcus gattii.Proc Natl Acad Sci U S A. 2025 Apr 29;122(17):e2503960122. doi: 10.1073/pnas.2503960122. Epub 2025 Apr 23. Proc Natl Acad Sci U S A. 2025. PMID: 40267138
-
Structural Characterization of CalS8, a TDP-α-D-Glucose Dehydrogenase Involved in Calicheamicin Aminodideoxypentose Biosynthesis.J Biol Chem. 2015 Oct 23;290(43):26249-58. doi: 10.1074/jbc.M115.673459. Epub 2015 Aug 3. J Biol Chem. 2015. PMID: 26240141 Free PMC article.
-
Analysis of nucleotide diphosphate sugar dehydrogenases reveals family and group-specific relationships.FEBS Open Bio. 2016 Jan 11;6(1):77-89. doi: 10.1002/2211-5463.12022. eCollection 2016 Jan. FEBS Open Bio. 2016. PMID: 27047744 Free PMC article.
References
-
- Feingold DS, Franzen JS. Pyridine nucleotide-linked four-electron transfer dehydrogenases. Trends in Biochemical Sciences. 1981;6:103–105.
-
- Trochon V, Mabilat C, Bertrand P, Legrand Y, Smadja-Joffe F, et al. Evidence of involvement of CD44 in endothelial cell proliferation, migration and angiogenesis in vitro. International Journal of Cancer. 1996;66:664–668. - PubMed
-
- Iozzo RV, Murdoch AD. Proteoglycans of the extracellular environment: clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB Journal. 1996;10:598–614. - PubMed
-
- Wight TN, Kinsella MG, Qwarnstrom EE. The role of proteoglycans in cell adhesion, migration and proliferation. Current Opinion in Cell Biology. 1992;4:793–801. - PubMed
-
- Jansen PL, Mulder GJ, Burchell B, Bock KW. New developments in glucuronidation research: report of a workshop on “glucuronidation, its role in health and disease”. Hepatology. 1992;15:532–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

