Contribution of the lipopolysaccharide to resistance of Shigella flexneri 2a to extreme acidity
- PMID: 21984920
- PMCID: PMC3184986
- DOI: 10.1371/journal.pone.0025557
Contribution of the lipopolysaccharide to resistance of Shigella flexneri 2a to extreme acidity
Abstract
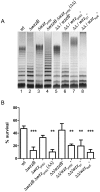
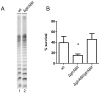
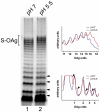
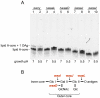
Shigella flexneri is endemic in most underdeveloped countries, causing diarrheal disease and dysentery among young children. In order to reach its target site, the colon, Shigella must overcome the acid environment of the stomach. Shigella is able to persist in this stressful environment and, because of this ability it can initiate infection following the ingestion of very small inocula. Thus, acid resistance is considered an important virulence trait of this bacterium. It has been reported that moderate acid conditions regulate the expression of numerous components of the bacterial envelope. Because the lipopolysaccharide (LPS) is the major component of the bacterial surface, here we have addressed the role of LPS in acid resistance of S. flexneri 2a. Defined deletion mutants in genes encoding proteins involved in the synthesis, assembly and length regulation of the LPS O antigen were constructed and assayed for resistance to pH 2.5 after adaptation to pH 5.5. The results showed that a mutant lacking O antigen was significantly more sensitive to extreme acid conditions than the wild type. Not only the presence of polymerized O antigen, but also a particular polymer length (S-OAg) was required for acid resistance. Glucosylation of the O antigen also contributed to this property. In addition, a moderate acidic pH induced changes in the composition of the lipid A domain of LPS. The main modification was the addition of phosphoethanolamine to the 1' phosphate of lipid A. This modification increased resistance of S. flexneri to extreme acid conditions, provide that O antigen was produced. Overall, the results of this work point out to an important role of LPS in resistance of Shigella flexneri to acid stress.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

