The dpsA gene of Streptomyces coelicolor: induction of expression from a single promoter in response to environmental stress or during development
- PMID: 21984935
- PMCID: PMC3184153
- DOI: 10.1371/journal.pone.0025593
The dpsA gene of Streptomyces coelicolor: induction of expression from a single promoter in response to environmental stress or during development
Erratum in
- PLoS One. 2011;6(11). doi:10.1371/annotation/8fedd9bb-d84e-4d7b-bc71-3321e652198d
Abstract
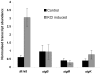
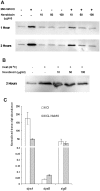
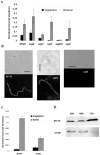
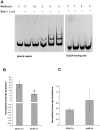
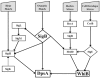
The DpsA protein plays a dual role in Streptomyces coelicolor, both as part of the stress response and contributing to nucleoid condensation during sporulation. Promoter mapping experiments indicated that dpsA is transcribed from a single, sigB-like dependent promoter. Expression studies implicate SigH and SigB as the sigma factors responsible for dpsA expression while the contribution of other SigB-like factors is indirect by means of controlling sigH expression. The promoter is massively induced in response to osmotic stress, in part due to its sensitivity to changes in DNA supercoiling. In addition, we determined that WhiB is required for dpsA expression, particularly during development. Gel retardation experiments revealed direct interaction between apoWhiB and the dpsA promoter region, providing the first evidence for a direct WhiB target in S. coelicolor.
Conflict of interest statement
Figures


References
-
- Hecker M, Pane-Farre J, Volker U. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–36. - PubMed
-
- Lee EJ, Karoonuthaisiri N, Kim HS, Park JH, Cha CJ, et al. A master regulator sigmaB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol Microbiol. 2005;57:1252–64. - PubMed
-
- Cho YH, Lee EJ, Ahn BE, Roe JH. SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol Microbiol. 2001;42:205–14. - PubMed
-
- Viollier PH, Kelemen GH, Dale GE, Nguyen KT, Buttner MJ, et al. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol Microbiol. 2003;47:699–714. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

