Genome-wide mapping of Sox6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by Sox6
- PMID: 21985497
- PMCID: PMC3239296
- DOI: 10.1186/1471-213X-11-59
Genome-wide mapping of Sox6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by Sox6
Abstract
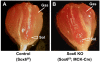
Background: Sox6 is a multi-faceted transcription factor involved in the terminal differentiation of many different cell types in vertebrates. It has been suggested that in mice as well as in zebrafish Sox6 plays a role in the terminal differentiation of skeletal muscle by suppressing transcription of slow fiber specific genes. In order to understand how Sox6 coordinately regulates the transcription of multiple fiber type specific genes during muscle development, we have performed ChIP-seq analyses to identify Sox6 target genes in mouse fetal myotubes and generated muscle-specific Sox6 knockout (KO) mice to determine the Sox6 null muscle phenotype in adult mice.
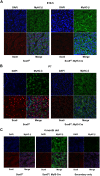
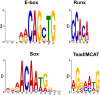
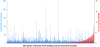
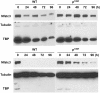
Results: We have identified 1,066 Sox6 binding sites using mouse fetal myotubes. The Sox6 binding sites were found to be associated with slow fiber-specific, cardiac, and embryonic isoform genes that are expressed in the sarcomere as well as transcription factor genes known to play roles in muscle development. The concurrently performed RNA polymerase II (Pol II) ChIP-seq analysis revealed that 84% of the Sox6 peak-associated genes exhibited little to no binding of Pol II, suggesting that the majority of the Sox6 target genes are transcriptionally inactive. These results indicate that Sox6 directly regulates terminal differentiation of muscle by affecting the expression of sarcomere protein genes as well as indirectly through influencing the expression of transcription factors relevant to muscle development. Gene expression profiling of Sox6 KO skeletal and cardiac muscle revealed a significant increase in the expression of the genes associated with Sox6 binding. In the absence of the Sox6 gene, there was dramatic upregulation of slow fiber-specific, cardiac, and embryonic isoform gene expression in Sox6 KO skeletal muscle and fetal isoform gene expression in Sox6 KO cardiac muscle, thus confirming the role Sox6 plays as a transcriptional suppressor in muscle development.
Conclusions: Our present data indicate that during development, Sox6 functions as a transcriptional suppressor of fiber type-specific and developmental isoform genes to promote functional specification of muscle which is critical for optimum muscle performance and health.
Figures
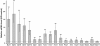
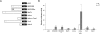
References
-
- Gunning P, Hardeman E. Multiple mechanisms regulate muscle fiber diversity. FASEB J. 1991;5:3064–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

