The stability of histone acetyltransferase general control non-derepressible (Gcn) 5 is regulated by Cullin4-RING E3 ubiquitin ligase
- PMID: 21987584
- PMCID: PMC3308846
- DOI: 10.1074/jbc.M111.290767
The stability of histone acetyltransferase general control non-derepressible (Gcn) 5 is regulated by Cullin4-RING E3 ubiquitin ligase
Abstract
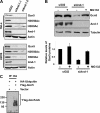
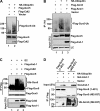
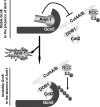
Histone acetyltransferases play important roles in the regulation of chromatin structure and gene transcription. As one of the most important histone acetyltransferases, general control non-derepressible (Gcn) 5 has been linked to diverse cellular processes and tumorigenesis as well. We have recently identified a functional link between Gcn5 and acidic nucleoplasmic DNA-binding protein 1 (And-1) that is elevated in multiple cancer cells and is essential for Gcn5 protein stability. However, the mechanism by which And-1 regulates Gcn5 protein stability remains unknown. Here we show that the ablation of Cullin4-RING E3 ubiquitin ligase (CRL4) leads to the stabilization of Gcn5 in cells with depleted And-1, and Cdc10-dependent transcript 2 (Cdt2) serves as a substrate receptor protein of CRL4. Overexpression of Cdt2 reduces the Gcn5 protein levels, and CRL(Cdt2) is sufficient to ubiquitinate Gcn5 both in vivo and in vitro. And-1 stabilizes Gcn5 by impairing the interaction between Gcn5 and CRL(Cdt2) and thereby preventing Gcn5 ubiquitination and degradation. The degradation of Gcn5 is not dependent on proliferating cell nuclear antigen, an important player involved in CRL(Cdt2)-mediated protein degradation. Thus, CRL(Cdt2) and And-1 play an essential role in the regulation of Gcn5 protein stability. This study provides us with the mechanistic basis to develop alternative approaches to inhibit Gcn5 activity for cancer therapy.
Figures




Similar articles
-
CRL4Cdt2 E3 ubiquitin ligase and proliferating cell nuclear antigen (PCNA) cooperate to degrade thymine DNA glycosylase in S phase.J Biol Chem. 2014 Aug 15;289(33):23056-23064. doi: 10.1074/jbc.M114.574210. Epub 2014 Jun 24. J Biol Chem. 2014. PMID: 24962565 Free PMC article.
-
CDK1-dependent inhibition of the E3 ubiquitin ligase CRL4CDT2 ensures robust transition from S Phase to Mitosis.J Biol Chem. 2015 Jan 2;290(1):556-67. doi: 10.1074/jbc.M114.614701. Epub 2014 Nov 19. J Biol Chem. 2015. PMID: 25411249 Free PMC article.
-
CRL4(CDT2) targets CHK1 for PCNA-independent destruction.Mol Cell Biol. 2013 Jan;33(2):213-26. doi: 10.1128/MCB.00847-12. Epub 2012 Oct 29. Mol Cell Biol. 2013. PMID: 23109433 Free PMC article.
-
CRL4Cdt2 Ubiquitin Ligase, A Genome Caretaker Controlled by Cdt2 Binding to PCNA and DNA.Genes (Basel). 2022 Jan 29;13(2):266. doi: 10.3390/genes13020266. Genes (Basel). 2022. PMID: 35205311 Free PMC article. Review.
-
CRL4Cdt2: Coupling Genome Stability to Ubiquitination.Trends Cell Biol. 2020 Apr;30(4):290-302. doi: 10.1016/j.tcb.2020.01.005. Epub 2020 Feb 7. Trends Cell Biol. 2020. PMID: 32044173 Review.
Cited by
-
The emerging role for Cullin 4 family of E3 ligases in tumorigenesis.Biochim Biophys Acta Rev Cancer. 2019 Jan;1871(1):138-159. doi: 10.1016/j.bbcan.2018.11.007. Epub 2018 Dec 30. Biochim Biophys Acta Rev Cancer. 2019. PMID: 30602127 Free PMC article. Review.
-
The stress phenotype makes cancer cells addicted to CDT2, a substrate receptor of the CRL4 ubiquitin ligase.Oncotarget. 2014 Aug 15;5(15):5992-6002. doi: 10.18632/oncotarget.2042. Oncotarget. 2014. PMID: 25115388 Free PMC article.
-
The CRL4DTL E3 ligase induces degradation of the DNA replication initiation factor TICRR/TRESLIN specifically during S phase.Nucleic Acids Res. 2021 Oct 11;49(18):10507-10523. doi: 10.1093/nar/gkab805. Nucleic Acids Res. 2021. PMID: 34534348 Free PMC article.
-
Overexpression of denticleless E3 ubiquitin protein ligase homolog (DTL) is related to poor outcome in gastric carcinoma.Oncotarget. 2015 Nov 3;6(34):36615-24. doi: 10.18632/oncotarget.5620. Oncotarget. 2015. PMID: 26472028 Free PMC article.
-
Ubiquitin proteasome system (UPS): a crucial determinant of the epigenetic landscape in cancer.Epigenomics. 2025 Jun;17(9):625-644. doi: 10.1080/17501911.2025.2501524. Epub 2025 May 8. Epigenomics. 2025. PMID: 40337853 Review.
References
-
- Shahbazian M. D., Grunstein M. (2007) Annu. Rev. Biochem. 76, 75–100 - PubMed
-
- Jones P. A., Baylin S. B. (2002) Nat. Rev. Genet. 3, 415–428 - PubMed
-
- Toyota M., Issa J. P. (2005) Semin. Oncol. 32, 521–530 - PubMed
-
- Brownell J. E., Zhou J., Ranalli T., Kobayashi R., Edmondson D. G., Roth S. Y., Allis C. D. (1996) Cell 84, 843–851 - PubMed
-
- Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D. (1996) Nature 383, 269–272 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

