A plant virus evolved by acquiring multiple nonconserved genes to extend its host range
- PMID: 21987809
- PMCID: PMC3198328
- DOI: 10.1073/pnas.1113227108
A plant virus evolved by acquiring multiple nonconserved genes to extend its host range
Abstract
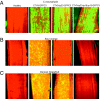
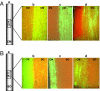
Viruses have evolved as combinations of genes whose products interact with cellular components to produce progeny virus throughout the plants. Some viral genes, particularly those that are involved in replication and assembly, tend to be relatively conserved, whereas other genes that have evolved for interactions with the specific host for movement and to counter host-defense systems tend to be less conserved. Closteroviridae encode 1-5 nonconserved ORFs. Citrus tristeza virus (CTV), a Closterovirus, possesses nonconserved p33, p18, and p13 genes that are expendable for systemic infection of the two laboratory hosts, Citrus macrophylla and Mexican lime. In this study, we show that the extended host range of CTV requires these nonconserved genes. The p33 gene was required to systemically infect sour orange and lemon trees, whereas either the p33 or the p18 gene was sufficient for systemic infection of grapefruit trees and the p33 or the p13 gene was sufficient for systemic infection of calamondin plants. Thus, these three genes are required for systemic infection of the full host range of CTV, but different genes were specific for different hosts. Remarkably, either of two genes was sufficient for infection of some citrus hybrids. These findings suggest that CTV acquired multiple nonconserved genes (p33, p18, and p13) and, as a result, gained the ability to interact with multiple hosts, thus extending its host range during the course of evolution. These results greatly extend the complexity of known virus-plant interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Lucas WJ. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology. 2006;344:169–184. - PubMed
-
- Lucas WJ, Gilbertson RL. Plasmodesmata in relation to viral movement within leaf tissue. Annu Rev Phytopathol. 1994;32:387–411.
-
- Wang HL, Wang Y, Giesman-Cookmeyer D, Lommel SA, Lucas WJ. Mutations in viral movement protein alter systemic infection and identify an intercellular barrier to entry into the phloem long-distance transport system. Virology. 1998;245:75–89. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

