Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis
- PMID: 21988170
- PMCID: PMC3315756
- DOI: 10.1089/scd.2011.0150
Conversion of human bone marrow-derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis
Abstract
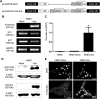
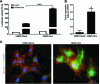
Tendons and ligaments (T/L) are dense connective tissues of mesodermal origin. During embryonic development, the tendon-specific cells descend from a sub-set of mesenchymal progenitors condensed in the syndetome, a dorsolateral domain of the sclerotome. These cells are defined by the expression of the transcription factor scleraxis (Scx), which regulates tendon formation and several other characteristic genes, such as collagen type I, decorin, fibromodulin, and tenomodulin (Tnmd). In contrast to other mesenchymal progenitors, the genealogy and biology of the tenogenic lineage is not yet fully understood due to the lack of simple and efficient protocols enabling generation of progenitors in vitro. Here, we investigated whether the expression of Scx can lead to the direct commitment of mesenchymal stem cells (MSCs) into tendon progenitors. First, MSC derived from human bone marrow (hMSC) were lentivirally transduced with FLAG-Scx cDNA to establish 2 clonal cell lines, hMSC-Scx and hMSC-Mock. Subsequent to Scx transduction, hMSC underwent cell morphology change and had significantly reduced proliferation and clonogenicity. Gene expression analysis demonstrated that collagen type I and several T/L-related proteoglycans were upregulated in hMSC-Scx cells. When stimulated toward 3 different mesenchymal lineages, hMSC-Scx cells failed to differentiate into chondrocytes and osteoblasts, whereas adipogenic differentiation still occurred. Lastly, we detected a remarkable upregulation of the T/L differentiation gene Tnmd in hMSC-Scx. From these results, we conclude that Scx delivery results in the direct programming of hMSC into tendon progenitors and that the newly generated hMSC-Scx cell line can be a powerful and useful tool in T/L research.
© Mary Ann Liebert, Inc.
Figures



Comment in
-
Are bone marrow mesenchymal stem cells-induced tenogenic progenitor cells identical to tendon progenitor cells?Stem Cells Dev. 2012 Apr 10;21(6):844-5. doi: 10.1089/scd.2011.0585. Epub 2012 Jan 17. Stem Cells Dev. 2012. PMID: 22150136 No abstract available.
References
-
- Kjaer M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol Rev. 2004;84:649–698. - PubMed
-
- Brent AE. Schweitzer R. Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113:235–248. - PubMed
-
- Tozer S. Duprez D. Tendon and ligament: development, repair and disease. Birth Defects Res C Embryo Today. 2005;75:226–236. - PubMed
-
- Schweitzer R. Chyung JH. Murtaugh LC. Brent AE. Rosen V. Olson EN. Lassar A. Tabin CJ. Analysis of the tendon cell fate using scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3866. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

