Identification of a gene for pyruvate-insensitive mitochondrial alternative oxidase expressed in the thermogenic appendices in Arum maculatum
- PMID: 21988877
- PMCID: PMC3327184
- DOI: 10.1104/pp.111.186932
Identification of a gene for pyruvate-insensitive mitochondrial alternative oxidase expressed in the thermogenic appendices in Arum maculatum
Abstract
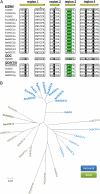
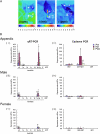
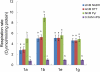
Heat production in thermogenic plants has been attributed to a large increase in the expression of the alternative oxidase (AOX). AOX acts as an alternative terminal oxidase in the mitochondrial respiratory chain, where it reduces molecular oxygen to water. In contrast to the mitochondrial terminal oxidase, cytochrome c oxidase, AOX is nonprotonmotive and thus allows the dramatic drop in free energy between ubiquinol and oxygen to be dissipated as heat. Using reverse transcription-polymerase chain reaction-based cloning, we reveal that, although at least seven cDNAs for AOX exist (AmAOX1a, -1b, -1c, -1d, -1e, -1f, and -1g) in Arum maculatum, the organ and developmental regulation for each is distinct. In particular, the expression of AmAOX1e transcripts appears to predominate in thermogenic appendices among the seven AmAOXs. Interestingly, the amino acid sequence of AmAOX1e indicates that the ENV element found in almost all other AOX sequences, including AmAOX1a, -1b, -1c, -1d, and -1f, is substituted by QNT. The existence of a QNT motif in AmAOX1e was confirmed by nano-liquid chromatography-tandem mass spectrometry analysis of mitochondrial proteins from thermogenic appendices. Further functional analyses with mitochondria prepared using a yeast heterologous expression system demonstrated that AmAOX1e is insensitive to stimulation by pyruvate. These data suggest that a QNT type of pyruvate-insensitive AOX, AmAOX1e, plays a crucial role in stage- and organ-specific heat production in the appendices of A. maculatum.
Figures



Similar articles
-
Degradation of mitochondrial alternative oxidase in the appendices of Arum maculatum.Biochem J. 2020 Sep 18;477(17):3417-3431. doi: 10.1042/BCJ20200515. Biochem J. 2020. PMID: 32856714 Free PMC article.
-
Interaction of purified alternative oxidase from thermogenic Arum maculatum with pyruvate.FEBS Lett. 2011 Jan 21;585(2):397-401. doi: 10.1016/j.febslet.2010.12.026. Epub 2010 Dec 25. FEBS Lett. 2011. PMID: 21187094
-
Different molecular bases underlie the mitochondrial respiratory activity in the homoeothermic spadices of Symplocarpus renifolius and the transiently thermogenic appendices of Arum maculatum.Biochem J. 2012 Jul 15;445(2):237-46. doi: 10.1042/BJ20111978. Biochem J. 2012. PMID: 22512685 Free PMC article.
-
[Alternative oxidase - never ending story].Postepy Biochem. 2016;62(2):138-148. Postepy Biochem. 2016. PMID: 28132465 Review. Polish.
-
Response of mitochondrial alternative oxidase (AOX) to light signals.Plant Signal Behav. 2011 Jan;6(1):55-8. doi: 10.4161/psb.6.1.14192. Epub 2011 Jan 1. Plant Signal Behav. 2011. PMID: 21270540 Free PMC article. Review.
Cited by
-
Metabolic interplay between cytosolic phosphoenolpyruvate carboxylase and mitochondrial alternative oxidase in thermogenic skunk cabbage, Symplocarpus renifolius.Plant Signal Behav. 2016 Nov;11(11):e1247138. doi: 10.1080/15592324.2016.1247138. Plant Signal Behav. 2016. PMID: 27739913 Free PMC article.
-
Structural and Biophysical Characterization of Purified Recombinant Arabidopsis thaliana's Alternative Oxidase 1A (rAtAOX1A): Interaction With Inhibitor(s) and Activator.Front Plant Sci. 2022 Jun 16;13:871208. doi: 10.3389/fpls.2022.871208. eCollection 2022. Front Plant Sci. 2022. PMID: 35783971 Free PMC article.
-
Thioredoxin o-mediated reduction of mitochondrial alternative oxidase in the thermogenic skunk cabbage Symplocarpus renifolius.J Biochem. 2019 Jan 1;165(1):57-65. doi: 10.1093/jb/mvy082. J Biochem. 2019. PMID: 30289493 Free PMC article.
-
Data analytic study of the homothermal maintenance mechanism of Skunk Cabbage: Capturing pre-equilibrium characteristics using extended poisson model.Biophys Physicobiol. 2018 Nov 6;15:235-250. doi: 10.2142/biophysico.15.0_235. eCollection 2018. Biophys Physicobiol. 2018. PMID: 30713824 Free PMC article.
-
Alternative Oxidase: From Molecule and Function to Future Inhibitors.ACS Omega. 2024 Mar 4;9(11):12478-12499. doi: 10.1021/acsomega.3c09339. eCollection 2024 Mar 19. ACS Omega. 2024. PMID: 38524433 Free PMC article. Review.
References
-
- Affourtit C, Albury MS, Crichton PG, Moore AL. (2002) Exploring the molecular nature of alternative oxidase regulation and catalysis. FEBS Lett 510: 121–126 - PubMed
-
- Affourtit C, Moore AL. (2004) Purification of the plant alternative oxidase from Arum maculatum: measurement, stability and metal requirement. Biochim Biophys Acta 1608: 181–189 - PubMed
-
- Albury MS, Affourtit C, Crichton PG, Moore AL. (2002) Structure of the plant alternative oxidase: site-directed mutagenesis provides new information on the active site and membrane topology. J Biol Chem 277: 1190–1194 - PubMed
-
- Albury MS, Elliott C, Moore AL. (2009) Towards a structural elucidation of the alternative oxidase in plants. Physiol Plant 137: 316–327 - PubMed
-
- Arnholdt-Schmitt B. (2009) Alternative oxidase (AOX) and stress tolerance: approaching a scientific hypothesis. Physiol Plant 137: 314–315 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

