Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene
- PMID: 21989056
- PMCID: PMC3276289
- DOI: 10.1093/hmg/ddr468
Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene
Abstract
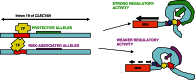
The chromosome 16p13 region has been associated with several autoimmune diseases, including type 1 diabetes (T1D) and multiple sclerosis (MS). CLEC16A has been reported as the most likely candidate gene in the region, since it contains the most disease-associated single-nucleotide polymorphisms (SNPs), as well as an imunoreceptor tyrosine-based activation motif. However, here we report that intron 19 of CLEC16A, containing the most autoimmune disease-associated SNPs, appears to behave as a regulatory sequence, affecting the expression of a neighbouring gene, DEXI. The CLEC16A alleles that are protective from T1D and MS are associated with increased expression of DEXI, and no other genes in the region, in two independent monocyte gene expression data sets. Critically, using chromosome conformation capture (3C), we identified physical proximity between the DEXI promoter region and intron 19 of CLEC16A, separated by a loop of >150 kb. In reciprocal experiments, a 20 kb fragment of intron 19 of CLEC16A, containing SNPs associated with T1D and MS, as well as with DEXI expression, interacted with the promotor region of DEXI but not with candidate DNA fragments containing other potential causal genes in the region, including CLEC16A. Intron 19 of CLEC16A is highly enriched for transcription-factor-binding events and markers associated with enhancer activity. Taken together, these data indicate that although the causal variants in the 16p13 region lie within CLEC16A, DEXI is an unappreciated autoimmune disease candidate gene, and illustrate the power of the 3C approach in progressing from genome-wide association studies results to candidate causal genes.
© The Author 2011. Published by Oxford University Press.
Figures





References
-
- Hakonarson H., Grant S.F., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Casalunovo T., Taback S.P., Frackelton E.C., et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. - PubMed
-
- Nischwitz S., Cepok S., Kroner A., Wolf C., Knop M., Muller-Sarnowski F., Pfister H., Rieckmann P., Hemmer B., Ising M., et al. More CLEC16A gene variants associated with multiple sclerosis. Acta Neurol. Scand. 2010;123:400–406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 079895/WT_/Wellcome Trust/United Kingdom
- G0800036/MRC_/Medical Research Council/United Kingdom
- 089989/WT_/Wellcome Trust/United Kingdom
- 076113/WT_/Wellcome Trust/United Kingdom
- BBS/E/B/0000C151/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0900951/MRC_/Medical Research Council/United Kingdom
- 076113/C/04/Z/WT_/Wellcome Trust/United Kingdom
- 091157/WT_/Wellcome Trust/United Kingdom
- 061858/WT_/Wellcome Trust/United Kingdom
- G0900729/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- RG/08/014/24067/BHF_/British Heart Foundation/United Kingdom
- G0800784/MRC_/Medical Research Council/United Kingdom
- 082549/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- 089989/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- BBS/E/B/0000M723/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

