Parameter estimation with bio-inspired meta-heuristic optimization: modeling the dynamics of endocytosis
- PMID: 21989196
- PMCID: PMC3271279
- DOI: 10.1186/1752-0509-5-159
Parameter estimation with bio-inspired meta-heuristic optimization: modeling the dynamics of endocytosis
Abstract
Background: We address the task of parameter estimation in models of the dynamics of biological systems based on ordinary differential equations (ODEs) from measured data, where the models are typically non-linear and have many parameters, the measurements are imperfect due to noise, and the studied system can often be only partially observed. A representative task is to estimate the parameters in a model of the dynamics of endocytosis, i.e., endosome maturation, reflected in a cut-out switch transition between the Rab5 and Rab7 domain protein concentrations, from experimental measurements of these concentrations. The general parameter estimation task and the specific instance considered here are challenging optimization problems, calling for the use of advanced meta-heuristic optimization methods, such as evolutionary or swarm-based methods.
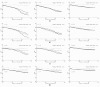
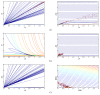
Results: We apply three global-search meta-heuristic algorithms for numerical optimization, i.e., differential ant-stigmergy algorithm (DASA), particle-swarm optimization (PSO), and differential evolution (DE), as well as a local-search derivative-based algorithm 717 (A717) to the task of estimating parameters in ODEs. We evaluate their performance on the considered representative task along a number of metrics, including the quality of reconstructing the system output and the complete dynamics, as well as the speed of convergence, both on real-experimental data and on artificial pseudo-experimental data with varying amounts of noise. We compare the four optimization methods under a range of observation scenarios, where data of different completeness and accuracy of interpretation are given as input.
Conclusions: Overall, the global meta-heuristic methods (DASA, PSO, and DE) clearly and significantly outperform the local derivative-based method (A717). Among the three meta-heuristics, differential evolution (DE) performs best in terms of the objective function, i.e., reconstructing the output, and in terms of convergence. These results hold for both real and artificial data, for all observability scenarios considered, and for all amounts of noise added to the artificial data. In sum, the meta-heuristic methods considered are suitable for estimating the parameters in the ODE model of the dynamics of endocytosis under a range of conditions: With the model and conditions being representative of parameter estimation tasks in ODE models of biochemical systems, our results clearly highlight the promise of bio-inspired meta-heuristic methods for parameter estimation in dynamic system models within system biology.
Figures








References
-
- Rice JA. Mathematical Statistics and Data Analysis. 3. Belmont, CA: Duxbury Press; 2007.
-
- Samaniego FJ. A Comparison of the Bayesian and Frequentist Approaches to Estimation. New York: Springer; 2010.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

