Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial
- PMID: 21990420
- PMCID: PMC3209811
- DOI: 10.1093/infdis/jir651
Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial
Abstract
Background: The HIV Prevention Trials Network (HPTN) 052 trial demonstrated that early initiation of antiretroviral therapy (ART) reduces human immunodeficiency virus (HIV) transmission from HIV-infected adults (index participants) to their HIV-uninfected sexual partners. We analyzed HIV from 38 index-partner pairs and 80 unrelated index participants (controls) to assess the linkage of seroconversion events.
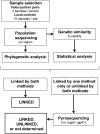
Methods: Linkage was assessed using phylogenetic analysis of HIV pol sequences and Bayesian analysis of genetic distances between pol sequences from index-partner pairs and controls. Selected samples were also analyzed using next-generation sequencing (env region).
Results: In 29 of the 38 (76.3%) cases analyzed, the index was the likely source of the partner's HIV infection (linked). In 7 cases (18.4%), the partner was most likely infected from a source other than the index participant (unlinked). In 2 cases (5.3%), linkage status could not be definitively established.
Conclusions: Nearly one-fifth of the seroconversion events in HPTN 052 were unlinked. The association of early ART and reduced HIV transmission was stronger when the analysis included only linked events. This underscores the importance of assessing the genetic linkage of HIV seroconversion events in HIV prevention studies involving serodiscordant couples.
Figures

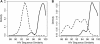

References
-
- Lam TT, Hon CC. Tang JW. Use of phylogenetics in the molecular epidemiology and evolutionary studies of viral infections. Crit Rev Clin Lab Sci. 2010;47:5–49. - PubMed
-
- Eshleman SH, Husnik M, Hudelson S, et al. Antiretroviral drug resistance, HIV-1 tropism, and HIV-1 subtype among men who have sex with men with recent HIV-1 infection. AIDS. 2007;21:1165–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01AI068619/AI/NIAID NIH HHS/United States
- U01 AI068619/AI/NIAID NIH HHS/United States
- U01AI068613/AI/NIAID NIH HHS/United States
- R01 DA024565/DA/NIDA NIH HHS/United States
- R37 AI029168/AI/NIAID NIH HHS/United States
- R01 AI029168/AI/NIAID NIH HHS/United States
- U01AI068617/AI/NIAID NIH HHS/United States
- U01 AI068613/AI/NIAID NIH HHS/United States
- UM1 AI068613/AI/NIAID NIH HHS/United States
- UM1AI068613/AI/NIAID NIH HHS/United States
- R01DA024565/DA/NIDA NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- U01 AI068617/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

