INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis
- PMID: 21990939
- PMCID: PMC3229140
- DOI: 10.1105/tpc.111.090944
INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis
Abstract
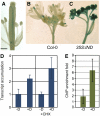
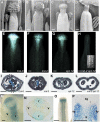
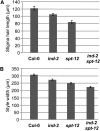
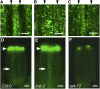
Structural organization of organs in multicellular organisms occurs through intricate patterning mechanisms that often involve complex interactions between transcription factors in regulatory networks. For example, INDEHISCENT (IND), a basic helix-loop-helix (bHLH) transcription factor, specifies formation of the narrow stripes of valve margin tissue, where Arabidopsis thaliana fruits open on maturity. Another bHLH transcription factor, SPATULA (SPT), is required for reproductive tissue development from carpel margins in the Arabidopsis gynoecium before fertilization. Previous studies have therefore assigned the function of SPT to early gynoecium stages and IND to later fruit stages of reproductive development. Here we report that these two transcription factors interact genetically and via protein-protein contact to mediate both gynoecium development and fruit opening. We show that IND directly and positively regulates the expression of SPT, and that spt mutants have partial defects in valve margin formation. Careful analysis of ind mutant gynoecia revealed slight defects in apical tissue formation, and combining mutations in IND and SPT dramatically enhanced both single-mutant phenotypes. Our data show that SPT and IND at least partially mediate their joint functions in gynoecium and fruit development by controlling auxin distribution and suggest that this occurs through cooperative binding to regulatory sequences in downstream target genes.
Figures



References
-
- Alvarez J., Smyth D.R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126: 2377–2386 - PubMed
-
- Alvarez J., Smyth D.R. (2002). CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 163: 17–41
-
- Balanzá V., Navarrete M., Trigueros M., Ferrándiz C. (2006). Patterning the female side of Arabidopsis: The importance of hormones. J. Exp. Bot. 57: 3457–3469 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

