Sortase A substrate specificity in GBS pilus 2a cell wall anchoring
- PMID: 21991306
- PMCID: PMC3186789
- DOI: 10.1371/journal.pone.0025300
Sortase A substrate specificity in GBS pilus 2a cell wall anchoring
Abstract
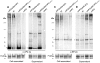
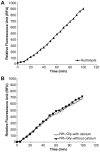
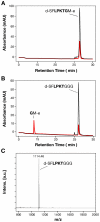
Streptococcus agalactiae, also referred to as Group B Streptococcus (GBS), is one of the most common causes of life-threatening bacterial infections in infants. In recent years cell surface pili have been identified in several Gram-positive bacteria, including GBS, as important virulence factors and promising vaccine candidates. In GBS, three structurally distinct types of pili have been discovered (pilus 1, 2a and 2b), whose structural subunits are assembled in high-molecular weight polymers by specific class C sortases. In addition, the highly conserved housekeeping sortase A (SrtA), whose main role is to link surface proteins to bacterial cell wall peptidoglycan by a transpeptidation reaction, is also involved in pili cell wall anchoring in many bacteria. Through in vivo mutagenesis, we demonstrate that the LPXTG sorting signal of the minor ancillary protein (AP2) is essential for pilus 2a anchoring. We successfully produced a highly purified recombinant SrtA (SrtA(ΔN40)) able to specifically hydrolyze the sorting signal of pilus 2a minor ancillary protein (AP2-2a) and catalyze in vitro the transpeptidation reaction between peptidoglycan analogues and the LPXTG motif, using both synthetic fluorescent peptides and recombinant proteins. By contrast, SrtA(ΔN40) does not catalyze the transpeptidation reaction with substrate-peptides mimicking sorting signals of the other pilus 2a subunits (the backbone protein and the major ancillary protein). Thus, our results add further insight into the proposed model of GBS pilus 2a assembly, in which SrtA is required for pili cell wall covalent attachment, acting exclusively on the minor accessory pilin, representing the terminal subunit located at the base of the pilus.
Conflict of interest statement
Figures


Similar articles
-
Structural differences between the Streptococcus agalactiae housekeeping and pilus-specific sortases: SrtA and SrtC1.PLoS One. 2011;6(8):e22995. doi: 10.1371/journal.pone.0022995. Epub 2011 Aug 30. PLoS One. 2011. PMID: 21912586 Free PMC article.
-
Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae.Infect Immun. 2008 Aug;76(8):3550-60. doi: 10.1128/IAI.01613-07. Epub 2008 Jun 9. Infect Immun. 2008. PMID: 18541657 Free PMC article.
-
In vitro reconstitution of sortase-catalyzed pilus polymerization reveals structural elements involved in pilin cross-linking.Proc Natl Acad Sci U S A. 2018 Jun 12;115(24):E5477-E5486. doi: 10.1073/pnas.1800954115. Epub 2018 May 29. Proc Natl Acad Sci U S A. 2018. PMID: 29844180 Free PMC article.
-
Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly.Protein Sci. 2017 Aug;26(8):1458-1473. doi: 10.1002/pro.3191. Epub 2017 May 15. Protein Sci. 2017. PMID: 28493331 Free PMC article. Review.
-
Anchoring of LPXTG-Like Proteins to the Gram-Positive Cell Wall Envelope.Curr Top Microbiol Immunol. 2017;404:159-175. doi: 10.1007/82_2016_8. Curr Top Microbiol Immunol. 2017. PMID: 27097813 Review.
Cited by
-
Understanding the molecular determinants driving the immunological specificity of the protective pilus 2a backbone protein of group B streptococcus.PLoS Comput Biol. 2013;9(6):e1003115. doi: 10.1371/journal.pcbi.1003115. Epub 2013 Jun 27. PLoS Comput Biol. 2013. PMID: 23825940 Free PMC article.
-
Surfaceome and Proteosurfaceome in Parietal Monoderm Bacteria: Focus on Protein Cell-Surface Display.Front Microbiol. 2018 Feb 14;9:100. doi: 10.3389/fmicb.2018.00100. eCollection 2018. Front Microbiol. 2018. PMID: 29491848 Free PMC article. Review.
-
LrpCBA pilus proteins of gut-dwelling Ligilactobacillus ruminis: crystallization and X-ray diffraction analysis.Acta Crystallogr F Struct Biol Commun. 2021 Aug 1;77(Pt 8):238-245. doi: 10.1107/S2053230X21007263. Epub 2021 Jul 28. Acta Crystallogr F Struct Biol Commun. 2021. PMID: 34341189 Free PMC article.
-
Structural basis for group B streptococcus pilus 1 sortases C regulation and specificity.PLoS One. 2012;7(11):e49048. doi: 10.1371/journal.pone.0049048. Epub 2012 Nov 8. PLoS One. 2012. PMID: 23145064 Free PMC article.
-
Pilin and sortase residues critical for endocarditis- and biofilm-associated pilus biogenesis in Enterococcus faecalis.J Bacteriol. 2013 Oct;195(19):4484-95. doi: 10.1128/JB.00451-13. Epub 2013 Aug 2. J Bacteriol. 2013. PMID: 23913319 Free PMC article.
References
-
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. - PubMed
-
- Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

