Nucleotide discrimination with DNA immobilized in the MspA nanopore
- PMID: 21991340
- PMCID: PMC3186796
- DOI: 10.1371/journal.pone.0025723
Nucleotide discrimination with DNA immobilized in the MspA nanopore
Abstract
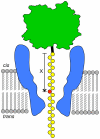
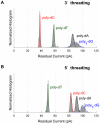
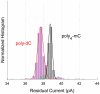
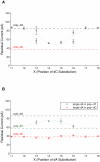
Nanopore sequencing has the potential to become a fast and low-cost DNA sequencing platform. An ionic current passing through a small pore would directly map the sequence of single stranded DNA (ssDNA) driven through the constriction. The pore protein, MspA, derived from Mycobacterium smegmatis, has a short and narrow channel constriction ideally suited for nanopore sequencing. To study MspA's ability to resolve nucleotides, we held ssDNA within the pore using a biotin-NeutrAvidin complex. We show that homopolymers of adenine, cytosine, thymine, and guanine in MspA exhibit much larger current differences than in α-hemolysin. Additionally, methylated cytosine is distinguishable from unmethylated cytosine. We establish that single nucleotide substitutions within homopolymer ssDNA can be detected when held in MspA's constriction. Using genomic single nucleotide polymorphisms, we demonstrate that single nucleotides within random DNA can be identified. Our results indicate that MspA has high signal-to-noise ratio and the single nucleotide sensitivity desired for nanopore sequencing devices.
Conflict of interest statement
Figures

References
-
- Bentley DR. Whole-genome re-sequencing. Curr Opin Genetics Dev. 2006;16:545–552. - PubMed
-
- Hirschhorn JN. Genomewide Association Studies – Illuminating Biologic Pathways. N Engl J Med. 2009;360:1699–1701. - PubMed
-
- Goldstein DB. Common Genetic Variation and Human Traits. N Engl J Med. 2009;360:1696–1698. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

