Engineering bispecificity into a single albumin-binding domain
- PMID: 21991353
- PMCID: PMC3185003
- DOI: 10.1371/journal.pone.0025791
Engineering bispecificity into a single albumin-binding domain
Abstract
Bispecific antibodies as well as non-immunoglobulin based bispecific affinity proteins are considered to have a very high potential in future biotherapeutic applications. In this study, we report on a novel approach for generation of extremely small bispecific proteins comprised of only a single structural domain. Binding to tumor necrosis factor-α (TNF-α) was engineered into an albumin-binding domain while still retaining the original affinity for albumin, resulting in a bispecific protein composed of merely 46 amino acids. By diversification of the non albumin-binding side of the three-helix bundle domain, followed by display of the resulting library on phage particles, bispecific single-domain proteins were isolated using selections with TNF-α as target. Moreover, based on the obtained sequences from the phage selection, a second-generation library was designed in order to further increase the affinity of the bispecific candidates. Staphylococcal surface display was employed for the affinity maturation, enabling efficient isolation of improved binders as well as multiparameter-based sortings with both TNF-α and albumin as targets in the same selection cycle. Isolated variants were sequenced and the binding to albumin and TNF-α was analyzed. This analysis revealed an affinity for TNF-α below 5 nM for the strongest binders. From the multiparameter sorting that simultaneously targeted TNF-α and albumin, several bispecific candidates were isolated with high affinity to both antigens, suggesting that cell display in combination with fluorescence activated cell sorting is a suitable technology for engineering of bispecificity. To our knowledge, the new binders represent the smallest engineered bispecific proteins reported so far. Possibilities and challenges as well as potential future applications of this novel strategy are discussed.
Conflict of interest statement
Figures
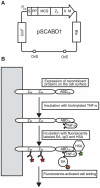


References
-
- Reichert JM. Monoclonal antibodies as innovative therapeutics. Curr Pharm Biotechnol. 2008;9:423–430. - PubMed
-
- Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. - PubMed
-
- Nygren PA. Alternative binding proteins: affibody binding proteins developed from a small three-helix bundle scaffold. FEBS J. 2008;275:2668–2676. - PubMed
-
- Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–1136. - PubMed
-
- Lofblom J, Frejd FY, Stahl S. Non-immunoglobulin based protein scaffolds. Curr Opin Biotechnol. 2011;22:1–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

